Abstract
Purpose
Extended spectrum β-lactamases (ESBLs) are cephalosporinases that confer resistance to a wide variety of oxyimino cephalosporins and create serious therapeutic problems. In addition, the quinolone resistance qnr genes are becoming increasingly prevalent in clinical isolates, some of which also produce ESBL. This study was designed to evaluate the occurrence and genotypic distribution of ESBL producing Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) as well as the prevalence and distribution of qnr genes in ESBL-producing isolates in a tertiary care hospital in Korea.
Materials and Methods
We tested a total of 111 ESBL-producing isolates of E. coli and K. pneumoniae, which were collected at Kyung Hee Medical Center from November 2006 to June 2008. ESBL production was determined by the Clinical and Laboratory Standards Institute (CLSI) ESBL confirmatory test. The cefotaxime and ceftazidime resistance of the ESBL-producers were transferred to azide-resistant E. coli J53 by conjugation. The presence and identity of ESBL and qnr genes were determined by polymerase chain reaction (PCR) and nucleotide sequencing.
Results
The prevalence of ESBLs was 17.7% (297/1,680) of E. coli and 26.5% (240/904) of K. pneumoniae in our hospital during the study periods. Of the 111 collected isolates, 69 isolates were E. coli and 42 isolates were K. pneumoniae. The most prevalent ESBL genotype was CTX-M15. Among the ESBL-producing isolates, 4 E. coli (5.8%) and 17 K. pneumoniae (40.5%) contained qnr genes. qnrB4 was the most frequent type in both E. coli and K. pneumoniae.
Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) are classified in the family Enterobacteriaceae. Species in these two genera are gram negative rods and frequently cause nosocomial and community-associated infections such as urinary tract infections, wound infections, and bacteremia. As narrow and extended-spectrum cephalosphorins are frequently used for treatment of infections by these species, resistance rates to these antimicrobial agents have also been increasing.1 Extended-spectrum β-lactamase (ESBL) producers are resistant to penicillins, narrow- and extended-spectrum cephalosporins, and aztreonam. These organisms are also frequently resistant to aminoglycosides, trimethoprim-sulfamethoxasole (TMP-SMZ), and quinolones. Therefore, the choice of antimicrobial agent to properly treat ESBL producers requires accurate identification. The ESBLs were first reported in 1983 and the prevalence and genotypes of ESBL-producing isolates have been reported in Korea since then.2-7
The first plasmid-mediated quinolone resistance-conferring gene (qnr) was reported in a K. pneumoniae clinical isolate from the USA in 1998.8 The qnr gene protects DNA gyrase against the effect of the quinolone.9 Recently, many studies have reported that the qnr gene is frequently encountered in ESBL-producing isolates.10-15
The distribution of ESBL genotypes has been found to vary according to the antimicrobial agents used in each hospital or local community.16,17 However, the distribution of genotypes of ESBL has not yet been reported in this hospital. Therefore, this study was designed to assess the prevalence and genotypes of ESBL-producing clinical isolates of E. coli and K. pneumoniae in Kyung Hee Medical Center as well as to evaluate the prevalence and genotypes of qnr genes in ESBL-producing isolates.
Nonduplicate ESBL-producing clinical isolates of E. coli and K. pneumoniae were randomly collected at Kyung Hee Medical Center from November 2006 to June 2008. All isolates were collected from inpatients. Conventional biochemical tests and MicroScan WalkAway 96 (Dade Behring, Sacramento, CA, USA) were used for bacterial identification.
For optimization of the PCR technique, well-characterized ESBL and qnr-positive strains were used as positive controls: K. pneumoniae SHV-12+,12 Shigella sonnei (S. sonnei) TEM-19+ (Korea Centers for Disease Control and Prevention, unpublished results), K. pneumoniae CTX-M3+,12 and S. sonnei CTX-M14+ (Korea Centers for Disease Control and Prevention, unpublished results) ; Enterobacter cloacae (E. cloacae) qnrA+, Citrobacter freundii qnrB+, and E. cloacae qnrS+.13
The phenotypic confirmatory test for ESBL production was performed by the disk diffusion method according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI).18
Antimicrobial susceptibilities were determined by agar dilution methods according to the recommendations of the CLSI.18 The tested antibiotics were amikacin (Boryung, Seoul, Korea), cefoxitin (Shinpoong, Seoul, Korea), cefepime (Boryung, Seoul, Korea), ceftazidime (LG Life Sciences, Seoul, Korea), cefotaxime (Handok, Seoul, Korea), aztreonam (Bristol-Myers Squibb, Princeton, NJ, USA), and ciprofloxacin (Bayer Vital GmbH, Berlin, Germany).
Conjugation experiments were carried out using the azide-resistant recipient strain E. coli J53 according to the method of Jacoby and Han.19 Each clinical and recipient strain was inoculated into brain heart infusion broth and incubated for 3 hours. 0.2 mL of donor culture solutions and 2.2 mL of recipient culture solutions were mixed in a tube and incubated at 37℃ for 1 hour. These mixed cultures were then inoculated on MacConkey agar supplemented with ceftazidime or cefotaxime (8 µg/mL, respectively) and 100 µg/mL of sodium azide (Sigma chemical Co., St. Louis, MO, USA). Transconjugants were selected after overnight incubation at 35℃ in ambient air. To confirm transference of resistance, transconjugants were subjected to the ESBL confirmation test and polymerase chain reaction (PCR) with primers listed in Table 1.
The plasmid deoxyribonucleic acid (DNA) was prepared using the Plasmid Purification Kit (SolGent Co., Daejeon, Korea) according to the manufacturer's recommendation. PCR amplification of the genes of ESBL and qnr genes were performed with primers listed in Table 1, as described previously.10,17,20 PCR products were analyzed by electrophoresis in a 2% agarose gel containing 0.5 µg/mL ethidium bromide at 120V for 40 min. The PCR products were purified with Agencourt Ampure (Beckman Coulter, Brea, CA, USA) and Biomek FX (Beckman Coulter, Brea, CA, USA) and sequenced on a 3730xl DNA analyzer (PE Applied Biosystems, Foster City, CA, USA). The nucleotide sequences were analyzed with software available from the National Center for Biotechnology Information BLAST service (http://www.ncbi.nlm.nih.gov/BLAST).
A total of 111 isolates, including 69 E. coli isolates and 42 K. pneumoniae isolates, were collected from the Kyung Hee Medical Center for analysis. These isolates were obtained from urine (64 isolates), sputum (27 isolates), blood (8 isolates), and other patient samples (12 isolates).
During the study period, the overall incidence of ESBL-producing isolates identified in the Kyung Hee Medical Center was 17.7% (297/1,680) of E. coli and 26.5% (240/904) of K. pneumoniae cases.
The resistance rates of E. coli and K. pneumoniae for amikacin, cefoxitin, cefepime, ceftazidime, cefotaxime, aztreonam, and ciprofloxacin are shown Table 2. Isolates containing qnr genes were highly resistant to ciprofloxacin (MIC ≥ 16 µg/mL) except for two K. pneumoniae isolates that were intermediately resistant (MIC 2 µg/mL). These isolates encoded qnrB4 and qnrS1, respectively.
Transconjugants were obtained for 20 strains (29.0%) of E. coli and 24 strains (57.1%) of K. pneumoniae using E. coli J53 Azr as the recipient strain, and tested positive in an ESBL confirmatory test and PCR test for blaSHV or blaCTX-M. One transconjugant displayed different results compared with the original isolate, which contained SHV5 and CTX-M15 while the transconjugant contained only CTX-M.
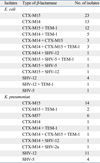
The distribution of the genotypes of ESBLs in this study is reported in Table 3. The most common types of ESBLs identified were CTX-M15 (n = 44, 63.8%) and CTX-M14 (n = 25, 36.2%) among E. coli isolates and CTX-M15 (n = 17, 40.5%) and SHV-12 (n = 12, 28.6%) among K. pneumoniae isolates. Genes encoding CTX-M57 (n = 6, 14.3%) were detected in K. pneumoniae isolates. Genes encoding TEM-type β-lactamases were detected in 22 of 69 E. coli isolates and 4 of 42 K. pneumoniae isolates, but all of them were the broad-spectrum TEM-1.
Among the ESBL producers, 5.8% (4/69) of E. coli and 40.5% (17/42) of K. pneumoniae contained qnr genes. Among the known qnr subtypes, only qnrB4 (n = 18, 85.7%) and qnrS1 (n = 3, 14.3%) were detected. The distribution of ESBL types in qnr containing isolates is shown in Table 4.
Although β-lactam antimicrobial agents are frequently used for the treatment of E. coli and K. pneumoniae infections, ESBL-producing isolates are becoming increasingly common. ESBL-producing isolates are of particular concern because these traits can be horizontally-transmitted to other isolates by plasmids and often cause nosocomial infections.21 The prevalence and genotype of ESBLs from clinical isolates vary according to the country and even hospital at which they are isolated from. For example, 3% (0-25%) of Enterobacteriaceae isolates from the USA were reported to be ESBL-producing strains22,23 while 0.1% of E. coli and 0.3% of K. pneumoniae isolates in Japan were ESBLs.24 Additionally, there have been several reports regarding the prevalence of ESBL-producing E. coli and K. pneumoniae in Korea, and previous reports presented that approximately 10% of E. coli and 25-30% of K. pneumoniae isolates produced ESBLs.5,6 ESBL-producing E. coli and K. pneumoniae isolates from Kyung Hee Medical Center were investigated, and among the 1,680 E. coli and 904 K. pneumoniae cases, a total of 297 E. coli isolates (17.7%) and 240 K. pneumoniae isolates (26.5%) were found to be ESBL-producing during the study periods. These results are similar to the ESBL incidence previously reported within Korea, although a slight increase in prevalence of ESBL-producing E. coli was detected.
Regarding the prevalence of the genotype of ESBLs in Korea, Pai25 reported that TEM-12 ESBL-producing E. coli and SHV-12 producing K. pneumoniae were the most common genotypes in the late 1990s. More recently, Hong, et al.26 reported that TEM-52 was the most prevalent in both E. coli and K. pneumoniae in 2001. In contrast, Kim, et al.,27 Rhoo, et al.,28 and Kang, et al.4 reported that CTX-M was most frequently produced in E. coli in 2005. Similarly, CTX-M14- and CTX-M15- producing E. coli were the most frequently encountered genotypes in this study. Surprisingly, TEM-52, the most frequently encountered genotype from previous reports, was not detected in this study. In the case of K. pneumoniae, SHV-12 accounted for 69% (27/39) of ESBLs in 199829 and 86% (31/36) of ESBLs in 200628 in Korea. However, in this study, CTX-M15 was the most prevalent (40.5%, 17/42), followed by SHV-12 (28.6%, 12/42). CTX-M3 or 15 types in E. coli and SHV-12 type in K. pneumoniae were predominant in most parts of Korea, but CTX-M3 and CTX-M14 types were most common in E. coli and K. pneumoniae, respectively, in a hospital in Gyeonggi in 2005.4 Our results were of a similar distribution to other hospitals in the different locations in Korea.
Interestingly, six isolates of K. pneumoniae expressed CTX-M57. CTX-M57 was first identified from Salmonella enterica in 2008.30 CTX-M57 shares 99% amino acid identity with CTX-M15 and differs from CTX-M15 by an A80V substitution. Though CTX-M occurred in 1.7% (9/520) of ESBLs in 2002,2 its prevalence within ESBLs increased to 4.3% (25/585) in 2005.28 This increase in prevalence of CTX-M is likely because gene acquisition of CTX-M or SHV is more effective than TEM in Enterobacteriaceae.17 In order to test for transference of β-lactam resistance, transconjugants were obtained from 29.0% (20/69) of E. coli and 57.1% (24/42) of K. pneumoniae tested using E. coli J53 Azr as the recipients strain, and represented positive results for the ESBL confirmatory test and tested positive in SHV or CTX-M by PCR. However, one transconjugant displayed different results compared with the original strain, which contained SHV5 and CTX-M15 while the transconjugant contained only CTX-M. This result suggests that SHV and CTX-M were not present on the same plasmid in the original isolate. There are several reports of isolates which contain both SHV and CTX-M, and these isolates express high resistance to not only ceftazidime but also ceftaxime. Therefore, a further investigation into whether SHV and CTX-M were present on same plasmid is required.
Quinolone resistance of Enterobactericeae is usually caused by various chromosomal mutations that alter the target enzymes, such as DNA gyrase, type IV topoisomerase, or active efflux systems. However, since the first plasmid-mediated quinolone resistance-conferring gene (qnr) was discovered in a K. pneumoniae isolate from USA, this gene has been reported worldwide,15,31,32 and is frequently encountered in ESBL-producing isolates.10,12,14 Recently, there have been some reports regarding the distribution of the qnr gene in Korea. Kim, et al.12 identified 44 isolates of ESBL-producing K. pneumoniae that tested positive for the qnrB gene, and 2 of these also tested positive for the qnrS gene. Shin, et al.33 reported that 5.6% (8/143) of E. coli and 55.9% (33/59) of K. pneumoniae isolates contained the qnrB gene among ciprofloxacin-resistant isolates. In this study, 5.8% (4/69) of E. coli and 33.3% (14/42) of K. pneumoniae had the qnrB4 gene and 7.1% (3/42) of K. pneumoniae had the qnrS1 gene. These results confirm previous reports regarding the high prevalence of the qnr gene, especially qnrB4, in K. pneumoniae. Interestingly, only one study13 has previously reported the identification of qnrS1 in Korea, which was found in a non-ESBL E. cloacae isolate. However, in this study, three isolates of K. pneumoniae contained the qnrS1. This could be an indication that the qnr gene has spread to several Enterobacteriaceae species. Two isolates of qnr-containing strains expressed intermediate susceptibility to ciprofloxacin. Previous analyses have reported that despite the cases that reported susceptibility to quinolone in the MIC test according to the CLSI guidelines, some isolates contained the qnr gene.33-36 The qnr-positive strain have been associated with only low-level resistance to fluoroquinolones in transconjugants,33 and most donor strains showed high levels of quinolone resistance with additional chromosomal mutations.9 Therefore, the presence of qnr may be contributed to the intensity and facilitation of quinolone resistance.9,33 In particular, because fluoroquinolones were widely used in Korea,37 the emergence and spread of qnr genes would contribute to the rapid development and spread of fluoroquinolone resistance.33 Therefore, we considered that early detection of qnr gene was very important for the prevention of the development and spread of this resistance.
In summary, CTX-M15 was the most frequently encountered in E. coli and K. pneumoniae isolated in the Kyung Hee Medical Center. In addition, new CTX-M57 was frequently detected in K. pneumoniae. Finally, a high prevalence of qnr genes on ESBL-producing strains was detected. β-lactam agents and quinolones should therefore be used cautiously in these species, and continuously monitored for resistance patterns.
Figures and Tables
Table 2
Susceptibility of ESBL-Producing E. coli and K. pneumoniae Isolates to 7 Antimicrobial Agents
References
1. Murray PR, Baron EJ, et al. Manual of Clinical Microbiology. 2007. 9th ed. Washington: American Society for Microbiology.
2. Bae IK, Woo GJ, Jeong SH, Park KO, Cho BK, Kim DM, et al. Prevalence of CTX-M-type extended-spectrum β-Lactamase-producing Esherichia coli and Klebsiella pneumoniae isolates in Korea. Korean J Clin Microbiol. 2004. 7:48–54.
3. Hong SG, Kim S, Jeong SH, Chang CL, Cho SR, Ahn JY, et al. Prevalence & diversity of extended-spectrum beta-Lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in Korea. Korean J Clin Microbiol. 2003. 6:149–155.
4. Kang JH, Bae IK, Kwon SB, Jeong SH, Lee J, Lee WG, et al. Prevalence of Ambler class A extended-spectrum beta-Lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in Korea. Korean J Clin Microbiol. 2005. 8:17–25.
5. Lee JH, Bae IK, Kwon SB, Jeong SH, Woo GJ, Lee J, et al. Prevalence of CTX-M-type extended-spectrum beta-Lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in Korea, 2003. Korean J Clin Microbiol. 2004. 7:111–118.
6. Park JH, Lee SH, Jeong SH, Kim BN, Kim KB, Yoon JD, et al. Characterization and prevalence of Escherichia coli and Klebsiella pneumoniae isolates producing an extended-spectrum beta-Lactamase from Korean hospitals. Korean J Lab Med. 2003. 23:18–24.
7. Song W, Kim JS, Kim MN, Kim EC, Park YJ, Yong D, et al. Occurrence and genotypic distributions of plasmid-mediated AmpC beta-Lactamase-producing Escherichia coli and Klebsiella pneumoniae in Korea. Korean J Lab Med. 2002. 22:410–416.
8. Jacoby GA, Chow N, Waites KB. Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother. 2003. 47:559–562.

9. Wang M, Sahm DF, Jacoby GA, Hooper DC. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother. 2004. 48:1295–1299.

10. Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007. 60:394–397.

11. Jones GL, Warren RE, Skidmore SJ, Davies VA, Gibreel T, Upton M. Prevalence and distribution of plasmid-mediated quinolone resistance genes in clinical isolates of Escherichia coli lacking extended-spectrum beta-lactamases. J Antimicrob Chemother. 2008. 62:1245–1251.

12. Kim MH, Sung JY, Park JW, Kwon GC, Koo SH. Coproduction of qnrB and armA from extended-spectrum beta-lactamase-producing Klebsiella pneumoniae. Korean J Lab Med. 2007. 27:428–436.

13. Park YJ, Yu JK, Lee S, Oh EJ, Woo GJ. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: a multicentre study from Korea. J Antimicrob Chemother. 2007. 60:868–871.

14. Wang A, Yang Y, Lu Q, Wang Y, Chen Y, Deng L, et al. Occurrence of qnr-positive clinical isolates in Klebsiella pneumoniae producing ESBL or AmpC-type beta-lactamase from five pediatric hospitals in China. FEMS Microbiol Lett. 2008. 283:112–116.

15. Wang A, Yang Y, Lu Q, Wang Y, Chen Y, Deng L, et al. Presence of qnr gene in Escherichia coli and Klebsiella pneumoniae resistant to ciprofloxacin isolated from pediatric patients in China. BMC Infect Dis. 2008. 8:68.

16. Bassetti M, Cruciani M, Righi E, Rebesco B, Fasce R, Costa A, et al. Antimicrobial use and resistance among Gram-negative bacilli in an Italian intensive care unit (ICU). J Chemother. 2006. 18:261–267.

17. Ko CS, Sung JY, Koo SH, Kwon GC, Shin SY, Park JW. Prevalence of extended-spectrum beta-lactamases in Escherichia coli and Klebsiella pneumoniae from Daejeon. Korean J Lab Med. 2007. 27:344–350.

18. CLSI. Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement, M100-S16. 2008. Wanye, PA: Clinical and Laboratory Standards Institute.
19. Jacoby GA, Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996. 34:908–911.

20. Bae IK, Jeong SH, Lee K, Yong D, Lee J, Hong SG, et al. Emergence of CTX-M12 and A Novel CTX-M Type Extended-Spectrum beta-Lactamaseproducing Klebsiella pneumoniae. Korean J Lab Med. 2006. 26:21–26.

21. Livermore DM. beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995. 8:557–584.

22. Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001. 14:933–951.

23. Jones RN, Pfaller MA, Doern GV, Erwin ME, Hollis RJ. Antimicrobial activity and spectrum investigation of eight broad-spectrum beta-lactam drugs: a 1997 surveillance trial in 102 medical centers in the United States. Cefepime Study Group. Diagn Microbiol Infect Dis. 1998. 30:215–228.

24. Yagi T, Kurokawa H, Shibata N, Shibayama K, Arakawa Y. A preliminary survey of extended-spectrum beta-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol Lett. 2000. 184:53–56.

25. Pai H. The characteristics of extended-spectrum beta-lactamases in Korean isolates of Enterobacteriaceae. Yonsei Med J. 1998. 39:514–519.

26. Hong SG, Kang M, Choi JR, Lee K, Chong Y, Kwon OH. Molecular characteristics of extended-spectrum beta-Lactamases in clinical isolates of Enterobacteriaceae. Korean J Clin Pathol. 2001. 21:495–504.
27. Kim J, Lim YM, Rheem I, Lee Y, Lee JC, Seol SY, et al. CTX-M and SHV-12 beta-lactamases are the most common extended-spectrum enzymes in clinical isolates of Escherichia coli and Klebsiella pneumoniae collected from 3 university hospitals within Korea. FEMS Microbiol Lett. 2005. 245:93–98.

28. Ryoo NH, Kim EC, Hong SG, Park YJ, Lee K, Bae IK, et al. Dissemination of SHV-12 and CTX-M-type extended-spectrum beta-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother. 2005. 56:698–702.

29. Kim J, Kwon Y, Pai H, Kim JW, Cho DT. Survey of Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J Clin Microbiol. 1998. 36:1446–1449.

30. Hopkins KL, Threlfall EJ, Karisik E, Wardle JK. Identification of novel plasmid-mediated extended-spectrum beta-lactamase CTX-M-57 in Salmonella enterica serovar Typhimurium. Int J Antimicrob Agents. 2008. 31:85–86.
31. Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005. 56:463–469.

32. Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006. 6:629–640.

33. Shin JH, Jung HJ, Lee JY, Kim HR, Lee JN, Chang CL. High rates of plasmid-mediated quinolone resistance QnrB variants among ciprofloxacin-resistant Escherichia coli and Klebsiella pneumoniae from urinary tract infections in Korea. Microb Drug Resist. 2008. 14:221–226.
34. Mammeri H, Van De Loo M, Poirel L, Martinez-Martinez L, Nordmann P. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother. 2005. 49:71–76.
35. Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci U S A. 2002. 99:5638–5642.

36. Wang M, Sahm DF, Jacoby GA, Zhang Y, Hooper DC. Activities of newer quinolones against Escherichia coli and Klebsiella pneumoniae containing the plasmid-mediated quinolone resistance determinant qnr. Antimicrob Agents Chemother. 2004. 48:1400–1401.

37. Shin JH, Kim HR, Lee HR, Chung JI, Min K, Moon CS, et al. Etiology and antimicrobial susceptibility of bacterial pathogens causing community-acquired urinary tract infection at a tertiary-care hospital. Korean J Clin Microbiol. 2005. 8:142–147.
38. Chong Y, Lee K, Okamoto R, Inoue M. Characteristics of Extended-spectrum beta-lactam hydrolyzing activity of Klebsiella pneumoniae and Escherichia coli strains isolated from clinical specimens. Korean J Infect Dis. 1997. 29:477–486.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download