Abstract
Purpose
Since November 2006, imipenem-resistant Acinetobacter baumannii isolates have increased in Kyung Hee University Hospital in Seoul, Korea. The purpose of this study was to determine the genetic basis and molecular epidemiology of outbreak isolates.
Materials and Methods
Forty-nine non-repetitive isolates of the 734 IRAB strains were investigated in order to determine their characteristics. The modified Hodge and the ethylenediaminetetraacetic acid (EDTA)-disk synergy test were performed for the screening of carbapenemase and metallo-β-lactamase production. Multiplex polymerase chain reaction (PCR) assays were performed for the detection of genes encoding for OXA-23-like, OXA-24-like, OXA-58-like and OXA-51-like carbapenemase. Pulsed-field gel electrophoresis (PFGE) was performed for strain identification.
Results
All isolates showed 100% resistance to ciprofloxacin and gentamicin, 97.9% resistance to cefepime, piperacillin/tazobactam, aztreonam, ceftazidime and piperacillin, 93.9% resistance to tobramycin and 57.1% resistance to amikacin. All of the 49 isolates (100%) showed positive results in the modified Hodge test and negative results in the EDTA-disk synergy test. They all (100%) possessed the encoding gene for an intrinsic OXA-51-like carbapenemase and an acquired OXA-23-like carbapenemase in the multiplex PCR assay. PFGE patterns revealed that all isolates were clonally related from A1 to A14.
Acinetobacter baumannii is a non-fermenting, aerobic Gram-negative bacillus that is distributed evenly in the hospital environment,1 and is an important opportunistic pathogen which causes nosocomial infections, especially in intensive care units. The known risk factors for A. baumannii colonization or infection include prolonged hospitalization, intensive care unit (ICU) admission, emergency surgical operation, total parenteral nutrition, invasive procedures, or previous broad-spectrum antibiotic use. A. baumannii causes a variety of nosocomial infections including septicemia, pneumonia, endocarditis, meningitis, wound infection and urinary tract infection.1 Carbapenems are the antibiotics of choice against multidrug resistant A. baumannii infections. However, the incidence of carbapenem-resistant A. baumannii is now being increasingly reported worldwide, thus causing serious therapeutic problems.2 Imipenem is one of the most effective carbapenems for the treatment for the infection caused by multidrug resistant A. baumannii. It causes good porin permeability of the outer membrane, and has affinity for penicillin-binding proteins, and stable β-lactamase.3 However, imipenem-resistant A. baumannii are being reported increasingly, ranging from 6.3% in 1999 to 13% in 2003 in Korea.4,5
Since November 2006, cases of imipenem-resistant A. baumannii have increased in Kyung Hee University Hospital in Seoul, Korea. The purpose of this study is to determine the genetic basis and molecular epidemiology of these outbreak isolates.
A total of 734 imipenem-resistant A. baumannii isolates were isolated from November 2006 to July 2007 at Kyung Hee University hospital in Seoul, Korea. The isolates were identified by conventional identification techniques and a MicroScan Walkaway 96 (Dade Behring, West Sacramento, CA, USA). Forty-nine (49) non-repetitive isolates of the 734 imipenem-resistant A. baumannii strains were investigated in order to determine their characteristics.
The testing was performed by two methods; One was minimum inhibitory concentrations (MIC) using a commercially prepared panel (MicroScan Walkaway 96) for blood specimens, and the other was the disk diffusion method for other specimens. The results were interpreted according to the guidelines of the Clinical Laboratory Standards Institute (CLSI).6
A modified Hodge test was performed to screen carbapenemase production. A suspension of Escherichia coli ATCC 25922, which was adjusted to the turbidity of the McFarland No. 0.5 tube was inoculated evenly on a Mueller-Hinton agar plate. Then, an imipenem disk (30 µg, BBL) was placed at the center of the plate. Test strains were streaked heavily from the edge of the disk to the periphery of the plate. The presence of a distorted inhibition zone after 16 to 18 hours of incubation at 35℃ was interpreted as a positive modified Hodge test. An ethylenediaminetetraacetic acid (EDTA)-disk synergy test was used for the screening of metallo-β-lactamase production. The test strains were suspended to the turbidity of the McFarland No. 0.5 tube and used to swab and inoculate a Mueller-Hinton agar plate. A 30 µg imipenem disk and a blank filter paper disk were placed 10 mm apart from edge to edge on the agar plate. Ten µL of 0.5 M EDTA solution was applied to the blank disk, which resulted in approximately a 1.5 mg/disk. After 16 to 18 hours of incubation at 35℃, the presence of an enlarged zone of inhibition was interpreted as EDTA-synergy test positive.7
A multiplex polymerase chain reaction (PCR) assay was performed for the detection of the carbapenem-resistant genes in the A. baumannii isolates according to the method described by Woodford, et al.8 These primers were combined with eight primers which were designed to amplify fragments of genes encoding for OXA-23-like, OXA-24-like, OXA-58-like and OXA-51-like carbapenemase. The amplification conditions were: initial denaturation at 94℃ for 5 minutes, 30 cycles of 94℃ for 25 seconds, 52℃ for 40 seconds, 72℃ for 50 seconds, and a final elongation at 72℃ for 6 minutes (Table 1).
The sample plugs were digested with 150 µL of ApaI reaction buffer containing 10 U of the ApaI restriction enzyme (New England Biolabs, Ipswich, MA, USA). DNA fragments were separated by electrophoresis in a 1% SeaKem gold agarose gel, using a 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.0) with CHEF Mapper® XA (Bio-Rad Laboratories, Hercules, CA, USA) at 14℃ and 6 V/cm. The electrophoresis was carried out by using alternating pulses at a 120° angle, with a 5-20 second pulse time gradient for 19 hours. Cluster analysis was performed using the unweighted pair group method with mathematical averaging (UPGMA). DNA relatedness was calculated using the band-based Dice coefficient with a tolerance setting of 1.0% band tolerance and 1.0% optimization setting for the entire profile. A similarity of less than 80% following dendogram analysis was considered to represent different PFGE types, while a similarity of greater than 80% was considered to represent PFGE subtypes.9
Data collected from the patient's medical records showed 30 isolates (61.2%) in the medical intensive care unit (MICU), 9 isolates (18.4%) in the Neurosurgical ICU (NICU), 5 isolates (10.2%) in the Surgical ICU (SICU), and 5 isolates (10.2%) in the general wards. Twenty-eight isolates (57.1%) were obtained from sputum samples. 7 isolates (14.3%) from noses, 7 isolates from blood samples and 4 isolates (8.2%) from wounds. In addition to those, 3 isolates were from urine, throat and other sites (Table 2).
All isolates showed 100% resistance to ciprofloxacin and gentamicin, 97.9% resistance to cefepime, piperacillin/tazobactam, aztreonam, ceftazidime and piperacillin, 93.9% resistance to tobramycin, and 57.1% resistance to amikacin (Table 3).
Among the 49 imipenem-resistant A. baumannii isolates, all 49 isolates (100%) showed positive results in the modified Hodge test and negative results in the EDTA-disk synergy test.
All 49 isolates (100%) possessed the encoding gene for an intrinsic OXA-51-like carbapenemase and an acquired OXA-23-like carbapenemase (Fig. 1).
All 49 imipenem-resistant A. baumannii isolates showed an identical band pattern and were classified as pulsotype A. Pulsotype A isolates were separated into fourteen subtypes, named subtypes A1 to A14. Sixteen isolates were subtype A7, three were A10, and two were A2 and A5. The other subtypes had one isolate each (Fig. 2).
A. baumannii is an organism which can colonize on the skin of healthy people. It has the ability to survive for a long time even in dry condition and the potential for airborne transmission.10,11 Because A. baumannii spreads easily through contamination of medical equipment used in patient monitoring or therapy, and from contamination by both patient and staff during handling of other environmental sources, these are difficult infections to control.1,12,13 Generally, carbapenems (imipenem and meropenem) are the most active agents for treating nosocomial infections caused by A. baumannii. However, the emergence of imipenem-resistant A. baumannii has become a worldwide problem which is causing serious therapeutic complications.2
Mechanism of A. baumannii resistance to carbapenems include the production of carbapenemase, the decreased outer-membrane permeability caused by the loss or reduced expression of porins, and the modification of penicillin-binding proteins. The most common resistance mechanism involves the acquisition of carbapenem-hydrolysing β-lactamase which belong to Ambler class B (metalloenzymes) and predominantly, Ambler class D (oxacillinase).14,15
Ambler class B is a powerful carbapenemase. It is classified as a metallo β-lactamase (MBL) because it requires Zn2+ for the efficient hydrolysis of β-lactams. Acquired MBLs are divided in to five types; IMP, VIM, SIM, SPM and GIM. However, only the first three of these types have been identified in A. baumannii. MBLs are susceptible to in vitro inhibition by EDTA, a chelater of divalent cations.14,15 IMP types have been reported worldwide, and VIM-2 and SIM-1 have been reported in A. baumannii isolates from South Korea.14,16,17
The Ambler class D carbapenemases of A. baumannii are divided into four phylogenetic subgroups; OXA-23-like (OXA-23, -27 and -49), OXA-24-like (OXA-24/40, -25, -26 and -72), OXA-58-like (OXA-59 and -96), and OXA-51-like carbapenemases. The OXA-23 subgroup was first reported in A. baumannii in Scotland in 1995 and was originally named ARI-1, but was renamed OXA-23.19-21 OXA-24 has been reported in Spain. OXA-40 in France, Spain and Portugal, OXA-23 in Brazil, French Polynesia, Spain, South Korea and England, and OXA-58 worldwide.22-29
The OXA-51-like subgroup may be intrinsic to A. baumannii, evidenced by its chromosomal location and its ubiquitous distribution among A. baumannii strains. However, it is not found in other Acinetobacter species. This enzyme may be involved in the expression of carbapenem-resistance under certain circumstances.30
Treatment for imipenem-resistant A. baumannii infections is sophisticated. Polymyxins (colinstimethate and polymyxin B) may be the only remaining therapeutic option. Colistin was used in the 1960s and 1970s, however was stopped due to severe adverse effects which included nephrotoxicity and neurotoxicity, as well as the emergence of safer alternative antimicrobials.3
The isolates of imipenem-resistant A. baumannii have increased abruptly since November 2006. Therefore, our infection control team was alarmed and started the surveillance program and the outbreak control. During the outbreak period, environmental contamination with IRAB was found on patient's bed, accessories of the mechanical ventilator and the surrounding environment. And the use of carbapenem was the significant risk factor for the acquisition of IRAB during December 2006 in multivariate analysis (p = 0.014). To control the outbreak, the hand washing after each patient contact, use of gloves and gowns, reinforcement of the environmental disinfection and the adequate management of the mechanical ventilator were rigorously enforced. The imipenem-resistant A. baumannii outbreak sustained for 9-months at our hospital from November 2006 to July 2007.
In 2005, the OXA-23-producing clones were reported in a University hospital, Busan, Korea that was the first report in Asia. Now we are certain that theses clones spread all over the nation.
In our study, all imipenem-resistant A. baumannii isolates were positive for carbapenemase production and negative for metallo-β-lactamase. They all possessed the encoding gene for an intrinsic OXA-51-like carbapenemase and an acquired OXA-23-like carbapenemase. All isolates analyzed had an identical band pattern, therefore this outbreak was caused by the spread of a clonally related epidemic.
The incidence of nosocomial infections due to multidrug resistant A. baumannii strains is increasing worldwide. These A. baumannii strains are rapidly adapting to the hospital environment, so that it is difficult to control the outbreaks. Early recognition of imipenem resistant A. baumannii clones is very important to prevent spreading within the hospital environment. Molecular typing for multidrug-resistant A. baumannii could be helpful in identification of a common source or cross contamination. This is an important step in tracing epidemiology of these strains.
Figures and Tables
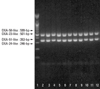 | Fig. 1Detection of genes encoding OXA carbapenemase by Multiplex PCR. Lane 1,100 bp Plus DNA Ladder (Bioneer, Daejeon, Korea); Lane 2-12, OXA-23-like and OXA-51-like. PCR, polymerase chain reaction. |
 | Fig. 2PFGE profiles of ApaI-digested genomic DNA from isolates of A. baumannii., PFGE, pulsed-field gel electrophoresis; A. baumannii, Acinetobacter baumannii. |
ACKNOWLEDGEMENTS
This research was supported by the Kyung Hee University Research Fund in 2008 (KHU-0599).
References
1. Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996. 9:148–165.

2. Jeong SH, Bae IK, Park KO, An YJ, Sohn SG, Jang SJ, et al. Outbreaks of imipenem-resistant Acinetobacter baumannii producing carbapenemases in Korea. J Microbiol. 2006. 44:423–431.
3. Murray CK, Hospenthal DR. Treatment of multidrug resistant Acinetobacter. Curr Opin Infect Dis. 2005. 18:502–506.

4. Lee K, Lee HS, Jang SJ, Park AJ, Lee MH, Song WK, et al. Antimicrobial resistance surveillance of bacteria in 1999 in Korea with a special reference to resistance of enterococci to vancomycin and gram-negative bacilli to third generation cephalosporin, imipenem, and fluoroquinolone. J Korean Med Sci. 2001. 16:262–270.

5. Hong SG, Yong DE, Lee KW, Kim EC, Lee WK, Jeung SH, et al. Antimicrobial resistance of clinically important bacteria isolated from hospitals located in representative provinces of Korea. Korean J Clin Microbiol. 2003. 6:29–36.
6. National Committee for Clinical Laboratory Standards. Document M100-S13. Performance standards for antimicrobial susceptibility testing: 13th informational supplement. 2003. Wayne, Pa: National Committee for Clinical Laboratory Standards.
7. Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified-Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001. 7:88–91.

8. Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006. 27:351–353.

9. Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005. 43:4328–4335.

10. McBride ME, Duncan WC, Knox JM. The Environment and the Microbial Ecology of Human Skin. Appl Environ Microbiol. 1977. 33:603–608.

11. Wagenvoort JH, Joosten EJ. An outbreak Acinetobacter baumannii that mimics MRSA in its environmental longevity. J Hosp Infect. 2002. 52:226–227.

12. Villers D, Espaze E, Coste-Burel M, Giauffret F, Ninin E, Nicolas F, et al. Nosocomial Acinetobacter baumannii infections: microbiological and clinical epidemiology. Ann Intern Med. 1998. 129:182–189.

13. Abbo A, Navon-Venezia S, Hammer-Muntz O, Krichali T, Siegman-Igra Y, Carmeli Y. Multidrug-resistant Acinetobacter baumannii. Emerg Infect Dis. 2005. 11:22–29.
14. Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii : mechanisms and epidemiology. Clin Microbiol Infect. 2006. 12:826–836.

15. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007. 20:440–458.
16. Yum JH, Yi K, Lee H, Yong D, Lee K, Kim JM, et al. Molecular characterization of metallo-beta-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the bla (VIM-2) gene cassettes. J Antimicrob Chemother. 2002. 49:837–840.

17. Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, et al. Novel acquired metallo-beta-lactamase gene, bla (SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005. 49:4485–4491.

18. Brown S, Amyes S. OXA (beta)-lactamases in Acinetobacter: the story so far. J Antimicrob Chemother. 2006. 57:1–3.
19. Paton R, Miles RS, Hood J, Amyes SG, Miles RS, Amyes SG. ARI 1: beta-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993. 2:81–87.
20. Scaife W, Young HK, Paton RH, Amyes SG. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995. 36:585–586.

21. Donald HM, Scaife W, Amyes SG, Young HK. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000. 44:196–199.

22. Bou G, Cerveró G, Domínguez MA, Quereda C, Martínez-Beltrán J. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of beta-lactamases. J Clin Microbiol. 2000. 38:3299–3305.

23. Héritier C, Poirel L, Aubert D, Nordmann P. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob Agents Chemother. 2003. 47:268–273.

24. Da Silva GJ, Quinteira S, Bértolo E, Sousa JC, Gallego L, Duarte A, et al. Long-term dissemination of an OXA-40 carbapenemase-producing Acinetobacter baumannii clone in the Iberian Peninsula. J Antimicrob Chemother. 2004. 54:255–258.

25. Dalla-Costa LM, Coelho JM, Souza HA, Castro ME, Stier CJ, Bragagnolo KL, et al. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J Clin Microbiol. 2003. 41:3403–3406.

26. Naas T, Levy M, Hirschauer C, Marchandin H, Nordmann P. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J Clin Microbiol. 2005. 43:4826–4829.

27. Marqué S, Poirel L, Héritier C, Brisse S, Blasco MD, Filip R, et al. Regional occurrence of plasmid-mediated carbapenem-hydrolyzing oxacillinase OXA-58 in Acinetobacter spp. in Europe. J Clin Microbiol. 2005. 43:4885–4888.

28. Poirel L, Marqué S, Héritier C, Segonds C, Chabanon G, Nordmann P. OXA-58, a novel class D {beta}-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005. 49:202–208.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download