Abstract
The developmental program of T helper and regulatory T cell lineage commitment is governed by both genetic and epigenetic mechanisms. The principal events, signaling pathways and the lineage determining factors involved have been extensively studied in the past ten years. Recent studies have elucidated the important role of chromatin remodeling and epigenetic changes for proper regulation of gene expression of lineage-specific cytokines. These include DNA methylation and histone modifications in epigenomic reprogramming during T helper cell development and effector T cell functions. This review discusses the basic epigenetic mechanisms and the role of transcription factors for the differential cytokine gene regulation in the T helper lymphocyte subsets.
The immune system consists of a complex array of cells that have developed to recognize and eradicate a wide variety of microorganisms while inducing tolerance against self-antigens and obnoxious antigens. The intriguing complexity of the immune system has been unraveled during the last two decades. Naive CD4 + T cells differentiate into T helper 1 (Th1), T helper 2 (Th2), T helper 17 (Th17) or regulatory T cells (Treg) depending on the microenvironment of antigenic stimulation by antigen presenting cells (Fig. 1). Different effector T cells are characterized by lineage-specific expression of cytokine genes. How immunity is influenced by signaling-mediated cytokine gene regulation program? What are the role of transcription factors, chromatin modifications and epigenetic mechanisms in the lineage commitment? Over the past ten years, considerable advances have been made to clarify the role of epigenetic regulation of gene expression patterns during the development of the pluripotent naive T helper cell into different effector subpopulations. In this article, we briefly discussed the mechanisms for CD4 + T helper cell development and lineage decisions. In addition, mechanisms of the epigenetic regulation of key cytokine genes in CD4 + T cell lineage commitment are also discussed.
T lymphocytes play an important role in the adaptive immune response to foreign pathogens and potentially to cancer cells. Naive T lymphocytes develop in the thymus and become activated in the periphery when they encounter foreign peptide antigens presented and processed by antigen presenting cells with the appropriate co-stimulatory signals. The proper differentiation and development into effector T cells (Th1, Th2, Th17 or Treg) in turn regulate immune responses to diverse antigens (including self- and non-self, pathogenic and non-pathogenic antigens) by producing lineage-specific effector cytokines. The lineage commitment of a naïve T cell into effector T cells is dependent on a series of events such as local cytokine environment and conditions of the TCR-MHC II interaction during antigen recognition, which is further directed by lineage-specific transcription factors that are capable of programming the expression of genes.1 The following subsections outline the general features of the T helper cell lineage development (Fig. 1).
Development of Th1 cells occurs in the presence of IL-12 and interferon gamma (IFN-γ) secreted by dendritic cells and macrophages in response to intracellular bacteria. IL-12 subsequently activates signal transducer and activator transcription4 (Stat4).2-4 The T-box transcription factor Tbet works as a "master regulator" of Th1 development,5,6 expression of which is induced by IFN-γ-stimulated Stat1 activation.7,8 Recent studies have shown the involvement of Jak3 and Stat5 in the development of Th1 cells.9 Th1 cells produce IL-2, IFN-γ and lymphotoxin, and promote cellular immune responses by activating macrophages or CD8 + cytotoxic T cells, but do not produce Th2 type cytokines such as IL-4 or IL-13.10,11
Differentiation and development towards the Th2 phenotype are initiated in the presence of IL-4 during T cell activation. Production of IL-4 in an early immune response directs the development of a Th2 response. Activated Th2 cells produce IL-4, IL-5, IL-13, and IgE, and activate mast cells and eosinophils,11-14 which play important roles in parasite removal and pathogenesis of Th2 type immune disorders such as asthma and atopic dermatitis. IL-4 receptor engagement leads to activation of Stat6, which in turn upregulates the transcription factor GATA3. GATA3 is regarded as the master regulator of Th2 development, a counterpart of T-bet in Th1 cells.15-19 Th2 cells produce IL-4, IL-5, IL-6, IL-10, and IL-13, and promote humoral immunity, but do not produce Th1 type cytokines such as IL-12 and IFN-γ. This exclusive pattern of cytokine production is mainly observed in fully committed Th1 and Th2 cells.10,11 C-maf, the basic-region leucine-zipper protein, is an another important transcription factor for Th2 development. It promotes IL-4 expression by directly binding to IL-4 promoter locus, and mice lacking c-maf are deficient in IL-4 production.20
Recent evidences have demonstrated that Th17 cells comprise a functionally distinct population and express high amounts of IL-17A, IL-17F and tumor necrosis factor-α (TNF-α). They can induce experimental autoimmune encephalomyelitis (EAE) upon passive transfer.21 IL-17 is a proinflammatory cytokine that mediates multiple chronic inflammatory response including angiogenesis, recruitment of inflammatory cells and induction of proinflammatory mediators by endothelial and epithelial tissues.25 A novel transcription factor, RORγt, is the central protein aiding Th17 development. STAT3 regulates RORγt expression.22 Both STAT3 and RORγt play a key role in IL-17 production and overexpression of either of them promotes IL-17 production.23 RORα also synergizes with RORγt to promote differentiation and function of Th17 cells.24
Within the pool of different effector T cells, the regulatory T cells (Tregs) are specialized subpopulation of T cells and play pivotal roles in maintaining immunological homeostasis. They modulate immune system through the induction of immune tolerance to self-antigens or unharmful antigens delivered through mucosal routes. Sakaguchi et al.26 demonstrated that Interleukin 2 receptor alpha (CD25) could serve as a phenotypic marker for CD4 + Tregs. Indeed, naturally occurring thymic derived CD4 + CD25 + Tregs have immunosuppressive function in nature, but Treg also expresses several other activation markers.27 Recent studies have identified the transcription factor forkhead box P3 (Foxp3) as a more exclusive intracellular marker for the identification of Tregs. Foxp3 also plays crucial roles for the development and functionality of CD4 + CD25 + Tregs. Ectopic expression of Foxp3 in T cells leads to generation of immunosuppressive regulatory T cells phenotype.28,29 Loss of Foxp3 function, both in mouse and human, results in absence of Tregs, leading to a phenotype with severe autoimmune disorders,30,31 known as scurfy mice, and IPEX (immunedysregulation, polyendocrinopathy, enteropathy, X-linked syndrome) in human. In addition to Treg, there are other subsets of antigen-induced or adaptive Tregs. CD4 + regulatory T cells of type 1 (Tr1) express high levels of IL-10 and moderate levels of IL-5, IFN-γ, and TGF-β. However, they do not produce IL-2 and IL-4.32,33 T helper 3 (Th3) regulatory T cells express high levels of TGF-β.34 Both types of induced Tregs equally suppress Th1- as well as Th2-mediated immune responses.
In eukaryotic cells, production of biologically active proteins is under sophisticated regulation at several points. Regulation of transcription initiation is the primary important step but the accessibility of transcription machinery mainly depends on the different chromatin structure such as euchromatin (open and accessible chromatin) versus heterochromatin (closed and condensed nonpermissive chromatin), which in turn mediates transcription levels and efficiency in each cell types. Modifications of DNA and DNA-binding histone molecules result in different chromatin structures. Histones are the basic components of a chromosome, in which the DNA helix (-147bp) is wrapped around four core histones (H2A, H2B, H3 and H4) to form the 'beads on a string structure', called nucleosomes that is then folded into higher order of dense chromatin fibres. Permissive epigenetic changes make structural alterations in chromatin organization for easy access of transcription machinery and allow active and selective gene transcription. Major epigenetic modifications such as DNA methylation and histone modifications, in concert with chromatin remodeling complexes, nuclear architecture and microRNAs, define the chromatin structure of a gene and its transcriptional activity. Although epigenetic marks are established early during development and differentiation, adaptations occur throughout life in response to intrinsic and environmental stimuli, which also mediate different cancers. The following subsections discuss the general principles of these epigenetic modifications (Fig. 2).
DNA itself can be modified via covalent addition of methyl groups on cytosines at CpG dinucleotides, catalyzed by enzymes known as DNA methyl transferases. About 40% of genes contain CpG-rich islands upstream from these transcriptional start sites, and up to 70% to 80% of all CpG dinucleotides in the genome are methylated.35 DNA methylation is involved in X-chromosome inactivation in females and DNA imprinting events, which result in monoallelic gene expression.36,37 DNA methylation is considered as a stable epigenetic mark and is maintatained in somatic cells by DNA methyltransferase I (DNMT I) with some cooperation of DNMT3a and DNMT3b.37,38 DNA methylation is usually involved in gene silencing and can repress gene expression by directly blocking the access of transcription regulatory factors to the target DNA.39 It can also recruit methyl-CpG-binding proteins (MECPs) in complex with histone deacetylases (HDACs) and co-repressor proteins like Sin3a and the multisubunit Nurd complex, repressing transcription in a methylation-dependent manner.40-44 Along with DNA methylation, the phenomenon of active DNA demethylation also exists as in the case of the IL-2 promoter which becomes demethylated within 20 minutes of stimulation.45,46 DNA methylation patterns in most cell types result from the balance of methylating (methyltransferases) and demethylating (demethylases) activities, and being targeted to specific genes by transcription factors acting downstream of signaling pathways which are yet to be unraveled.38
Various types of posttranslational histone modifications include acetylation, methylation, ubiquitylation, phosphorylation and sumoylation.47 Most posttranslational modifications of histones are found in the N- and C-terminal tails, and some are associated with active chromatin structure and some act as repressive marks. The combinatorial pattern of modifications on histones is interpreted by the cell as an epigenetic code from the genome to the cellular machinery, commonly termed as the histone code hypothesis.
The most predominant and best understood covalent modification is acetylation of histones. During specific biological processes, selected lysines such as lysine (K) 9 and 14 are acetylated by enzymes called histone acetyltransferases (HATs) which catalyze the attachment of acetyl groups. Histone deacetylases (HDACs) are enzymes which help in removal of these acetyl groups and act in concert with the HATs to maintain a steady state balance.48-50 The H4 tail also has a prominent role in compaction of DNA fibers. Acetylation of H4K16 and loss or reduction of linker histones result in a decondensed chromatin fibre.38,51
Histone methylation on the other hand is a more stable form of modification than acetylation52 and is among the least understood histone modifications. The most heavily methylated histone is H3 followed by H4.48 Histone methylation occurs on lysine (K) residues 4, 9, 27 and 36 on H3 and on position 20 on H4, and might provide an ideal epigenetic mark for more long-term maintenance of chromatin states.53 Histone methyltransferases (HMTs) are the enzymes that regulate the site-specific methylation of lysine residues; for example, K9 and K4 in amino terminus of histone H3. Standard HMTs contain evolutionarily conserved 130 amino acid SET-domain, thereby stimulating some of the biological processes such as gene activation or repression depending on the properties of binding proteins.54
Histone hyperacetylation along with di- or tri-methylation of K4 of H3 (H3K4me2, K4me3) is generally associated with chromatin decondensation, accessibility of DNA to binding proteins and increased transcriptional activity, wheras histone hypoacetylation and H3K9me2/3 and H3K27me3 constitute repressive marks and contribute to chromatin condensation and transcriptional repression.39, 55-58
Control of gene transcription in developmental stage-specific or signal-dependent manner is one of the most important mechanisms to maintain the intrinsic functional properties of each cell. Cis-regulatory DNA regions act as switches to control the on/off state of genes in particular cell types.59 Transcription initiation occurs at core promoters and is further regulated by enhancers, silencers, insulators and locus control regions (LCRs).60 Cis-regulatory modules are composed of multiple transcription factor binding sites61 and are highly conserved in vertebrates.62 A number of these sequences has been shown to be transcriptional regulatory regions. Enhancers contain binding sites for transcription factors required for maximal transcriptional efficiency. Silencers often bind to transcription factors having repressive effect on transcription. On the other hand, insulators block the communication between enhancers or silencers and promoter regions and protect genes from the influence of neighboring gene segments and chromatin.62 Locus control regions are gene segments containing enhancers, silencers and insulators, and they enhance the transcription of specific genes in a copy-dependent manner. Their regulatory action is orientation-independent over long distances. Locus control regions have been identified in several genomic regions including the Th2 locus, β-globin locus and the gene cluster, known as the human growth hormone locus.63,64 Different chromatin architecture, especially in the cis-regulatory DNA regions, mediates the accessibility of transcriptosome complex, which in turn regulates transcription levels and efficiency in lineage or developmental specific gene expression patterns.
Cytokines such as IL-4 and IFN-γ control T helper cell differentiation and are critical regulator for adaptive immune responses. Naïve CD4 + T cells upon stimulation show basal expression levels of IL-4 and IFN-γ, implying that regulatory elements of both cytokines are in a poised state.65 During development of naïve CD4 + T cells into cytokine-producing effector cells in response to antigen stimulation, T helper cells differentiate into distinct Th1 or Th2 cell lineages, characterized by differential expression of cytokine genes. Naïve T helper cells differentiate to Th1 cells in the presence of IL-12 and IFN-γ, while differentiate to Th2 cells in the presence of IL-4. Th1 cells produce IL-2, IFN-γ and lymphotoxin and promote cellular immune responses by activating macrophages or CD8 + cytotoxic T cells, however, do not produce IL-4 and IL-13. In contrast, Th2 cells produce IL-4, IL-5, IL-6, IL-10 and IL-13 and promote humoral immunity, but do not produce IFN-γ. These mutually exclusive patterns of cytokine production are mainly observed in fully committed Th1 and Th2 cells in vivo and in vitro. Recent studies on exclusive cytokine expression profile of Th1 (IFN-γ) and Th2 (IL-4) demonstrated opposing epigenetic regulation depending on the direction of polarization.1,66,67
In naïve T cells, most of CpG dinucleotides at regulatory elements are demethylated. Differentiation to Th1 cells showed similar level of DNA methylation to that of naïve T cells, but some of the regulatory elements showed increased demethylation, while Th2 differentiation was associated with substantial overall methylation.68 Unlike the IL-4 promoter region, methylation of IFN-γ promoter in T cells has been controversial. From the detailed quantitative analysis of six CpGs in promoter regions, it has been confirmed that IFN-γ promoter becomes methylated during Th2 cell development. In contrast, the promoter region as well as transcribed region becomes demethylated in naïve and differentiated Th1 cells,69 demonstrating that various changes in DNA methylation at the IFN-γ locus occur during Th1 and Th2 cell development. In addition, there are increased levels of H3 and H4 acetylation, H3K4 dimethylation and DNase I hypersensitive sites at the IFN-γ locus and complete loss of H3K27 methylation, which is the representative mark of repression, in the IFN-γ regulatory elements in Th1 cells.68,70-72 While differentiated Th2 cells show loss of permissive marks, repressive H3K27 trimethylation appears along with increased level of CpG methylation throughout the IFN-γ locus.68,73 Th1-specific changes of permissive histone modification are acquired by STAT4 and T-bet. STAT4 binds to the promoter and other elements like CNS-22 and this leads to increased levels of permissive histone modifications. Moreover, BRG1, known as a core factor of chromatin remodeling complexes, also interacts with STAT4, and this induces the IFN-γ mRNA by promoting permissive histone modificaiotns.74 T-bet, a master regulator of Th1 development, also induces Th1 development by increasing expression level of IFN-γ in STAT4-dependent or -independent manner.75,76 T-bet alone could bind to mehtylated DNA in promoter region, and it has recently been reported that T-bet can interact with jumonji-domain-containing protein histone demethylase 3 (JMJD3) and histone methyltransferase SET7, resulting in increased levels of permissive histone modification in Th1 cells.77,78
Important regulatory elements in IL-4-IL-13 locus show highly demethylated DNA level during Th2 development and this correlates with result from 5-azacytidine treatment or MBD2-deficient T cells.79-81 In mouse Th2-cytokine locus, many DNase I hypersensitive sites are correlated with the enhanced IL-4 expression in Th2 cells as well as increased permissive histone modification. Permissive H3 and H4 acetylation and H3K4 dimethylation are more enriched throughout the locus, and repressive H3K27 trimethylation is diminished at the same time.66,82-85 As naïve T cells differentiate into Th2 cells, increased expression of GATA-3 is necessary for changes in epigenetic modifications within the IL-4 locus.17,75,82 However, the exact mechanism by which GATA3 induces epigenetic modifications remains to be explored. STAT6 also induces Th2 differentiation through its binding to various regulatory elements.83 Thus, epigenetic mechanisms in the fully committed Th1 or Th2 lineage population from naïve T cells shows clear differences in DNase I hypersensitivity, DNA methylation and histone modifications.1,79,86 Much facts are still unknown as for the detailed mechanisms by which factors play crucial role in epigenetic regulation and the in vivo functional relevance between the regulatory elements and the epigenetic changes therein.
Unlike IFN-γ and IL-4, IL-10 is produced in both recently differentiated primary Th1 and Th2 cells even though Th2 cells produce much higher level (5-7 times depending on Th2 phenotype with or without re-stimulation) than Th1 cells.87,88 This is very unique phenomenon compared to exclusive expression profile of IFN-γ and IL-4 in Th1 and Th2 cells, respectively. As naïve T cells differentiate to Th1 and Th2 cells, Th1 cells slowly lose their ability to express IL-10 after 1 week of differentiation, while Th2 cells show increased IL-10 production and maintain high IL-10 levels (Lee et al., unpublished result). These results suggest a dynamic chromatin remodeling on IL-10 locus during T cell differentiation. Although the biological function of IL-10 in immune system has extensively been studied for decades, little information is available on the molecular mechanism of its transcriptional regulation, especially at the chromatin level. Attempts to identify cis-regulatory elements have been made using DNase I HSS mapping. In our previous work, we described the HSS and identified 6 HSS in CD4 + T cells.89 Later, Wang et al.90 showed that the CNS + 6.45 region is a constitutive DNase I hypersensitive site, and that JunB and c-Jun bind to this region specifically in Th2 cells. Jones and Flavell87 reported that the CNS-9 region is transcriptionally active in both a Th1 clone (AE7) and a Th2 clone (D10). However, there are no reports about detailed epigenetic regulation mechanism in the primary Th1 and Th2 cell system.
Chang et al. showed that IL-10 locus in Th1 cells displayed little acetylation of histone H4 at 3 weeks of skewing in Th1 conditions, while Th2 cells within 1 week of skewing showed enhanced histone H4 acetylaton throughout the locus. The level of acetylation of H4 increased more in 3 weeks of skewing to Th2 cells, therefore, they concluded that IL-10 gene showed epigenetic imprinting in Th2 cells, but not in Th1 cells.91 Our own work showed that Th2 cells have elevated acetylation (on histone H3 at Lys9 and Lys14) and methylation (on histone H3 at Lys4) at all the CNS sites compared to Th1 cells (Lee et al., unpublished result). This change of acetylation level could partly be mediated by GATA-3, because Shoemaker et al.92 previously reported that GATA-3 induces acetylation of H3 and H4 of IL-10 gene in promoter region, introns and 3' downstream region. There is also a possibility of involvement of Th1-specific active silencing during Th1 development in the IL-10 locus. Th1 cells express higher IL-10 levels than naïve CD4 + T cells upon stimulation, although much lower than Th2 cells. Indeed, Th1 cells showed higher binding of the active chromatin markers than naïve T cells, but less than that of Th2 cells. Both Th1 and Th2 cells might be programmed to express IL-10. Unlike Th2 cells, Th1-specific recruitment of a silencing complex to cis-regulatory elements, including promoter or other regulatory elements could lead to lower IL-10 expression in Th1 cells.
Not much is known about the epigenetic and regulatory mechanisms which govern the Th17-cell differentiation and development. IL-17A and IL-17F are typically known representative cytokines of in vivo and in vitro differentiated Th17 cells.93 Permissive H3 acetylation is involved at the promoter of IL17a and IL17f genes, and STAT3 is the known regulator of this histone modification.94,95 RORγt, the master regulator in Th17 cells, binds to regulatroy locus CNS region rather than promoter.24 However, there are still much unknown facts about the epigenetic regulation in Th17 cells and the factors involved in the regulatory mechanism governing IL17a and IL17f expression.
Foxp3 is the master regulator for the function of Treg and could be induced in the peripheral CD4 + 25- T cells.96 Like other genes, DNA methylation has crucial role in maintaining Foxp3 gene expression. Treatment of DNA demethylating agent, 5-azacytidine, significantly affects Foxp3 expression during induced Treg generation.97-99 Treatment of TGF-β together with TCR stimulation induces Treg (iTreg) generation by increasing Foxp3 production. Recently, regulatory elements involved in TGF-β mediated iTreg generation has been identified as TGF-β sensor that is responsible for the binding of transcription factors such as NFAT and SMAD3.100 However, little is known how those transcriptional regulators are recruited into proximal promoter region of Foxp3 under induced Treg generation conditions and what are their exact role as activators or repressors.96 In addition, extensive investigations are needed to elucidate the role of epigenetic reprogramming and histone modifying factors in generation of Treg cells.
The lineage commitment of T helper cells is crucial in controlling immune homeostasis. T helper cell differentiation is tightly regulated by transcription of crucial target genes through epigenetic modification with combinatorial network of transcription factors. So far, many studies have revealed unique epigenetic marks in distinct cells, and differentiation stages determine expression kinetics of target genes. These include DNA methylation, histone modification and recruitment of transcription factors in the genomic locus of target genes. However, more extensive investigations are needed to understand the detailed processes of epigenetic regulation. First, even though the outcome of epigenetic changes leads to different levels of transcription, there is no fine mapping of how each epigenetic mark acts in a transcriptional process. Second, the mechanisms of how specific epigenetic modifications are modulated by specific transcription factors should be revealed to further strengthen the relationship between epigenetic marks and modulators such as transcription factors, chromatin modifiers and other proteins. Third, the mechanisms of how epigenetic marks of epigenetic memory in lineage commitment are specifically established in different differentiation conditions should be explored more in detail. Finally, there are more layers of mechanisms for proper regulation of genes during development and differentiation such as higher order chromatin remodeling and three dimensional conformation of chromosomal locus in nucleus. With recent development of systemic approach and cutting edge technologies in gene regulation study, we are hopeful to be able to better understand the diversity of T helper cell development and the epigenetic mechanisms associated.
Figures and Tables
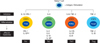 | Fig. 1Diversification of naïve CD4 + T helper cells into various effector T helper cell lineages. Upon antigenic stimulation, naïve CD4 + T cells can be differentiated into diverse T helper cell subsets like Th1, Th2, Th17 and Treg. From in vitro and in vivo studies, instructive cytokines and transcription factors which are specific for each effector lineage were identified. In the early initiation stage, the unique signal transducer and activator of transcription (STAT) is activated by the environmental cytokine signals which leads to induction of lineage master regulators. Transcription factors from each distinct subset activate and control the various downstream genes and this mechanism is further enhanced and stabilizes the lineage commitment with epigenetic modification by specific stimuli and the action of transcription factors. As a result, effector cytokines and modulators can be released from each CD4 + T helper cell subsets and these further regulate immune responses accordingly to antigens. |
 | Fig. 2Epigenetic modifications in T helper cells. Histone modifications and DNA methylation are two major epigenetic mechanisms which governs gene expression in mammalian cells. First, DNA methylation is detected from the 5-cytosine of CpG dinucleotides or CpG islands which is generally associated with the mechanism of gene silencing. Posttranslational modification of histone molecules occurs mainly at H4 or H4 tails. Histone tails are easily modified by the external stimuli and consist of various types of modifications such as acetylation, methylation, phosphorylation, and sumoylation. Among these, acetylation and methylation on lysine residues are well known representative modifications during T helper cell differentiation. Level of acetylation and lysine 4 methylation of Histone H3 and H4 are generally linked with active and accessible state of gene regulation while methylation of lysine 9 and 27 are well known marks of silent or inactive gene regulation. |
ACKNOWLEDGEMENTS
Our work described in this paper was supported by grants from the Regional Technology Innovation Program of the MOCIE (RTI05-01-01), the Korea Healthcare technology R&D Project, Ministry for Health, Welfare and (A080588-5), a Systems Biology Infrastructure Establishment Grant and by the Center for Distributed Sensor Network at GIST.
References
1. Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003. 4:616–623.

2. Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of TH1 CD4 + T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993. 260:547–549.

3. Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996. 382:174–177.
4. Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996. 382:171–174.

5. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000. 100:655–669.

6. Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002. 295:338–342.

7. Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat Immunol. 2002. 3:549–557.

8. Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001. 98:15137–15142.
9. Shi M, Lin TH, Appell KC, Berg LJ. Janus-kinase-3-dependent signals induce chromatin remodeling at the lfng locus during T helper 1 cell differentiation. Immunity. 2008. 28:763–773.

10. Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996. 383:787–793.

12. Romagnani S. Lymphokine production by human T cells in disease states. Annu Rev Immunol. 1994. 12:227–257.

13. Sher A, Coffman RL. Regulation of immunity to parasites by T cells and T cell-derived cytokines. Annu Rev Immunol. 1992. 10:385–409.
14. Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th2-like helper effectors. J Immunol. 1990. 145:3796–3806.
15. Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996. 4:313–319.
16. Kurata H, Lee HJ, O'Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999. 11:677–688.
17. Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O'Garra A, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000. 192:105–115.

18. Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996. 380:630–633.
19. Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996. 380:627–630.

20. Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999. 10:745–751.

21. Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005. 201:233–240.

22. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006. 126:1121–1133.

23. Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007. 282:9358–9363.

24. Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008. 28:29–39.

25. Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008. 28:454–467.

26. Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995. 155:1151–1164.
27. Sakaguchi S. Regulatory T cells: key controllers of immunologic self-tolerance. Cell. 2000. 101:455–458.
28. Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol. 2005. 6:331–337.

29. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003. 299:1057–1061.

30. Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001. 27:68–73.

31. Gambineri E, Torgerson TR, Ochs HD. Immune dysregulation, polyendocrinopathy, enteropathy, and X-linked inheritance (IPEX), a syndrome of systemic autoimmunity caused by mutations of FOXP3, a critical regulator of T-cell homeostasis. Curr Opin Rheumatol. 2003. 15:430–435.

32. Roncarolo MG, Bacchetta R, Bordignon C, Narula S, Levings MK. Type 1 T regulatory cells. Immunol Rev. 2001. 182:68–79.

33. Roncarolo MG, Gregori S, Battaglia M, Bacchetta R, Fleischhauer K, Levings MK. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev. 2006. 212:28–50.

34. Weiner HL. Oral tolerance: immune mechanisms and the generation of Th3-type TGF-beta-secreting regulatory cells. Microbes Infect. 2001. 3:947–954.

36. Illingworth R, Kerr A, Desousa D, Jørgensen H, Ellis P, Stalker J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008. 6:e22.

37. Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007. 213:384–390.

39. Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002. 3:662–673.

40. Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001. 15:827–832.
41. Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998. 19:187–191.

42. Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998. 393:386–388.
43. Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999. 23:58–61.

44. Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999. 13:1924–1935.

45. Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003. 4:235–240.

48. Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. J Genet Genomics. 2009. 36:75–88.

49. Kuo Min-Hao, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998. 20:615–626.

50. Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001. 70:81–120.
51. Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008. 381:816–825.
52. Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007. 447:407–412.

54. Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001. 11:266–273.

55. Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004. 11:1037–1043.

58. Sims RJ 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006. 20:2779–2786.

59. Strähle U, Rastegar S. Conserved non-coding sequences and transcriptional regulation. Brain Res Bull. 2008. 75:225–230.
60. Janson PC, Winerdal ME, Winqvist O. At the crossroads of T helper lineage commitment-Epigenetics points the way. Biochim Biophys Acta. 2008. 12. 30. [Epub ahead of print].
61. Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004. 116:247–257.

62. Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000. 16:369–372.
63. Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003. 19:145–153.

64. West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005. 14:R101–R111.
65. Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001. 14:205–215.
66. Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T (H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002. 3:643–651.
67. Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002. 2:933–944.

68. Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007. 8:732–742.

69. Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006. 25:2443–2452.
70. Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998. 9:765–775.
71. Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004. 279:4802–4810.
72. Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, et al. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004. 101:12622–12627.
73. Chang S, Aune TM. Dynamic changes in histone-methylation 'marks' across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007. 8:723–731.
74. Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med. 2006. 203:1493–1505.
75. Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002. 169:647–650.
76. Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001. 292:1907–1910.

77. Tong Y, Aune T, Boothby M. T-bet antagonizes mSin3a recruitment and transactivates a fully methylated IFN-gamma promoter via a conserved T-box half-site. Proc Natl Acad Sci U S A. 2005. 102:2034–2039.
78. Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008. 22:2980–2993.

79. Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006. 24:369–379.

80. Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, et al. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998. 9:229–237.
81. Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002. 10:81–91.
82. Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004. 21:865–876.

83. Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc Natl Acad Sci U S A. 2004. 101:16010–16015.

84. Baguet A, Bix M. Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc Natl Acad Sci U S A. 2004. 101:11410–11415.

85. Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005. 280:31470–31477.

86. Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009. 9:91–105.

87. Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J Immunol. 2005. 175:7437–7446.

88. Lee CG, Kang KH, So JS, Kwon HK, Son JS, Song MK, et al. A distal cis-regulatory element, CNS-9, controls NFAT1 and IRF4-mediated IL-10 gene activation in T helper cells. Mol Immunol. 2008. 46:613–621.

89. Im SH, Hueber A, Monticelli S, Kang KH, Rao A. Chromatin-level regulation of the IL 10 gene in T cells. J Biol Chem. 2004. 279:46818–46825.
90. Wang ZY, Sato H, Kusam S, Sehra S, Toney LM, Dent AL. Regulation of IL-10 gene expression in Th2 Cells by Jun proteins. J Immunol. 2005. 174:2098–2105.

91. Chang HD, Helbig C, Tykocinski L, Kreher S, Koeck J, Niesner U, et al. Expression of IL-10 in Th memory lymphocytes is conditional on IL-12 or IL-4, unless the IL-10 gene is imprinted by GATA-3. Eur J Immunol. 2007. 37:807–817.

92. Shoemaker J, Saraiva M, O'Garra A. GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J Immunol. 2006. 176:3470–3479.

93. Ivanov II, Zhou L, Littman DR. Transcriptional regulation of Th17 cell differentiation. Semin Immunol. 2007. 19:409–417.

94. Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007. 282:5969–5972.

95. Wei L, Laurence A, Elias KM, O'Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007. 282:34605–34610.
96. Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007. 5:e38.

97. Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007. 204:1543–1551.

98. Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008. 38:1654–1663.
99. Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009. 182:259–273.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download