Abstract
Purpose
Enterobacter sakazakii (E. sakazakii) infections are an important cause of life-threatening meningitis, septicemia, and necrotizing enterocolitis in infants. Dried infant formula milk is an important vehicle for E. sakazakii infection. E. sakazakii was isolated in Korea from dried infant formula milk. Although E. sakazakii infection of infants may occur in Korea, its prevalence has not yet been documented. Therefore, we determined the prevalence of E. sakazakii and documented symptoms.
Materials and Methods
Between March and October 2006, 1,146 stool samples were collected from patients at Uijeongbu St. Mary's Hospital. Each fecal swab was dissolved in 10 mL of buffered peptone solution, and enriched culture was streaked onto Druggan-Forsythe-Iversen (DFI) agar. Presumptive E. sakazakii colonies that exhibited a blue-green color during culture on DFI medium were selected. The identity of colonies that developed yellow pigment during culture on TSA was determined using the Vitek system and PCR.
Enterobacter sakazakii (E. sakazakii) is a bacterium that belongs to the family of Enterobacteriaceae. It was initially referred to as yellow-pigmented Enterobacter cloacae. In 1980, Farmer et al.1 reclassified it as E. sakazakii, based on DNA-DNA hybridization, biochemical characteristics, antibiotic susceptibility patterns, and presence of yellow-pigmented colonies.
E. sakazakii infection is an important cause of life-threatening meningitis (complicated by ventriculitis, brain abscess, cerebral infarction, and cyst formation), septicemia, and necrotizing enterocolitis in infants.2,3 Mortality rates of 33-80% were reported for infected patients.2 There are few reports of E. sakazakii infection of adults, which is not usually life threatening.3-5
E. sakazakii has been isolated from stool and urine.6-8 Isolation from stools was demonstrated in infants aged ≤ 18 weeks.9 This organism has also been detected in a wide range of foods, including cheese, meat, vegetables, grains, herbs, and spices.10-12 Most reports are concerned with the presence of the organism in dried infant formula milk.10-13 Dried infant formula milk is an important vehicle of E. sakazakii infection.14 In Korea, E. sakazakii was isolated from 3 of 45 dried infant formula milk samples.15 Although E. sakazakii infection of infants may occur in Korea, its prevalence has not yet been determined. Therefore, we determined the prevalence of E. sakazakii in stool samples of hospital patients in Korea and documented the symptoms of those whose stool samples were positive for E. sakazakii.
To isolate E. sakazakii, 1,146 stool samples were collected from March to October 2006 at Uijeongbu St. Mary's Hospital.
Each fecal swab from each sample was dissolved in 10 mL of buffered peptone solution (Oxoid, Basingstroke, England) and incubated at 37℃ for 24 hours. One mL of preenriched buffered peptone solution was added to 10 mL of enterbacter enrichment broth (Oxoid) and incubated at 37℃ for 24 hours. The enriched culture was streaked onto Druggan-Gorsythe-iversen (DFI) medium (Oxoid) and incubated at 37℃ for 24 hours. Up to five presumptive E. sakazakii colonies that exhibited a blue-green color during culture on DFI medium were selected for culture on TSA at 25℃ for 72 hours. The identities of colonies that developed yellow pigment during culture on TSA were determined using a Vitek GNI card (bio Meriux, Marcyl' Etoile, France) and PCR. E. sakazakii ATCC 29004, E. sakazakii ATCC 29544, and E. sakazakii ATCC 51329 were used as positive controls.
Bacteria were grown on TSA at 37℃ for 24 hours. One loop of biomass was scraped off the agar, suspended in 500 µL of sterile distilled water, and boiled for 10 minutes. After centrifugation at 1200 × g for 10 minutes at 4℃, the supernatants were used as templates for PCR. To detect the sequence of the 16S rRNA gene, E. sakazakii strains were amplified using PCR assays with Esakf (5' GCT YTG CTG ACG AGT GGC GG 3') and Esakr (5' ATC TCT GCA GGA TTC TCT GG 3'), details of which have previously been reported.7
The following conditions were used for the thermal cycler (PTC-100; MJ Research, Watertown, MA, USA): initial denaturation for 2 minutes at 94℃, 29 cycles of denaturation at 94℃ for 30 seconds, annealing at 60℃ for 60 seconds, extension at 72℃ for 90 seconds, and a final cycle at 72℃ for 5 minutes. The products were then sequenced using a modified Sanger method, a Big-dye kit (Applied Biosystems), and an ABI 3,730 capillary DNA analyzer (Applied Biosystems, Foster City, CA, USA).
The antibiotic susceptibility of E. sakazakii was determined using the Vitek system and GNS cards. The following antibiotics were tested: amikacin (AN), ampicillin (AM), ampicillin/sulbactam (SAM), aztreonam (AZ), cefazolin (CZ), cefepime (FEP), cefoxitin (CFX), ciprofloxacin (CIP), gentamicin (GM), imipenem (IPM), piperacillin/tazobactam (TAZ), and trimethoprime-sulfamethoxazole (SXT).
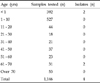
Four E. sakazakii strains were isolated from 1,146 stool samples of Korean patients. Details of stool samples and isolated strains are shown in Table 1. E. sakazakii was isolated from 2 of 392 infants less than one year of age. Among adults, 2 E. sakazakii strains were isolated from 61 - 70-year-old age group. The 4 clinical isolates and 3 ATCC strains were tested for antibiotic resistance. Strains isolated from babies less than one year of age were resistant to ampicillin and cefazolin, whereas isolates from adults were susceptible to all antibiotics tested. The clinical characteristics of the 4 cases were as follows.
A 3-month-old baby who exhibited symptoms of poor nutrition and abdominal distention was hospitalized, and a clinical diagnosis of gastroenteritis and meningitis was made. The white blood cell (WBC) count of cerebrospinal fluid (CSF) was 16 per mm3 and consisted of lymphocytes (82%), monocytes (12%), and neutrophils (6%). CSF agglutination tests for Streptococcus agalactiae, S. pnemoniae, Neisseria meningitidis, and H. influenza type B were negative. Blood and CSF cultures were negative. Intravenous therapy with amoxicillin/clavulanate and tobramycin was initiated and the patient's condition gradually improved after 10 days of hospitalization. Rotavirus antigen and E. sakazakii were isolated from stool specimens on day 7 of hospitalization.
A 20 days-old baby presented with fever. He was hospitalized with a clinical diagnosis of sepsis and meningitis. The patient's physical condition on admission was normal except for the presence of jaundice. Intravenous therapy with ampicillin/sulbactam and tobramycin was initiated, and the patient's condition gradually improved after 5 days of hospitalization. A diagnostic lumbar puncture revealed that CSF had a total protein concentration of 81 mg/dL, a glucose concentration of 62 mg/dL, and a WBC of 2 per mm3. CSF agglutination tests for Streptococcus agalactiae, S. pnemoniae, Neisseria meningitidis, and H. influenza type B were negative. Blood and CSF cultures were negative. E. sakazakii was isolated from stool specimens after 4 days of hospitalization.
A 64 years-old male patient presented with right hemiplegia and thymic cyst rupture. E. sakazakii was isolated from stool specimens on day 30 of hospitalization. No sign or symptom indicative of E. sakazakii infection was observed during hospitalization.
A 72 years-old male patient was diagnosed with intracranial hemorrhage. E. sakazakii was isolated from stool specimens on day 30 of hospitalization. No sign or symptom indicative of E. sakazakii infection was observed during hospitalization.
The biochemical and antibiotic resistance characteristics of the isolates are presented in Table 2. Three of the 4 isolates produced yellow pigmentation during culture on TSA. Moreover, all 4 isolates exhibited a blue-green color during culture on DFI agar, which confirmed the constitutive expression of α-glucosidase (data not shown). Three of the Vitek GNI biochemical profiles of isolates identified as E. sakazakii had identification values of 96 - 99% and that of one strain was 82% (S2, Table 2). There were several point mutations in the sequences of the strains (Table 3).
We isolated four E. sakazakii strains from 1,146 stool samples. Clinical outbreaks of E. sakazakii infection in neonatal care units in various parts of the world have been reported.16,17 However, isolation of E. sakazakii from humans in Korea has never been documented before.
All four isolates developed blue-green colonies on DFI agar, confirming the constitutive expression of α-glucosidase. However, α-glucosidase activity is not restricted to E. sakazakii.18
E. sakazakii is naturally resistant to all macrolides, lincomycin, clindamycin, streptogramins, rifampicin, fusidic acid, and fosfomycin.18 It is, however, susceptible to some antibiotics, including tetracyclines, aminoglycosides, numerous α-lactams, chloramphenicol, antifolates, and quinolones.19 Our results showed that strains of E. sakazakii isolated from adults were sensitive to ampicillin and cefazolin. However, the strains of E. sakazakii isolated from infants were resistant to ampicillin and cefazolin. These observations are different from those of Mustjens et al., who showed that the resistance to ampicillin is likely due to exclusion of E. sakazakii.20 We speculate that specific virulence factor could explain the differences in susceptibility between the strains isolated from infants and those from adults. But, further study is needed to confirm the differences in antibiotic susceptibilities.
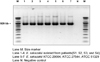
Several studies on the molecular characterization of E. sakazakii have been conducted to identify phylogenetic relationships and suitable PCR primers.19,21-23 The 16S rRNA gene has widely been used for sequence analysis because it is highly conserved and ubiquitous. Amplification of the 16S rRNA gene from the E. sakazakii isolates using the Esakf and Esakr primer pair produced PCR products with 929 bp long (Fig. 1). These primers are very specific for E. sakazakii.19 Molecular methods based on 16S rRNA gene sequencing are well established and are routinely used for characterization and identification of bacteria.24,25 The 16S rRNA gene contains variable and hypervariable regions. The variable regions have been used to discriminate between species and genera. Variability in the E. sakazakii 16S rRNA gene has also been observed. Lehner et al.19 detected 2 phylogenetically distinct lineages in the 16S rRNA gene, and Hassan et al.26 observed sequence differences between E. sakazakii and other Enterobacteriaceae within hypervariable regions. We compared the 16S rRNA sequences of the 4 isolates with those of the three ATCC strains. The sequence alignments of the 16S rRNA PCR products of the 4 isolates and the 3 E. sakazakii ATCC strains had 99% similarity. However, we detected several point mutations in the sequences (Table 3). The present result together with previous study19 indicate that the E. sakazakii 16S rRNA gene has at least 4 "hot spots" and may contain several point mutations. A few cases of E. sakazakii infection in adult have been reported, and younger than 4 years old infants and children are high risk groups compared with adults.10
In this study, infection signs of E. sakazakii were encountered in only 2 patients, however, not seen in adults. This result shows that E. sakazakii infection is more risky in infants than in adults. Powdered infant formula is both the vehicle and the direct/indirect source of 50 - 80% cases of diseases caused by E. sakazakii.14 Therefore, we suggest that dried infant formula milk in the present study could be considered as the possible sources of E. sakazakii isolated from infants, although we were unable to prove our hypothesis. Finally, our study shows that E. sakazakii infection does occur in Korea.
In conclusion, this study is the first to report the isolation of E. sakazakii from stool samples of Korean patients and indicates that continuous surveillance is needed to monitor E. sakazakii infection.
Figures and Tables
References
1. Farmer JJ III, Asbury MA, Hickman FW, Brenner DJ. Enterobacteriaceae Study Group. Enterobacter sakazakii: a new species of "Enterobacteriaceae" isolated from clinical specimens. Int J Syst Bacteriol. 1980. 30:569–584.

2. Lai KK. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore). 2001. 80:113–122.

3. Nazarowec-White M, Farber JM. Enterobacter sakazakii: a review. Int J Food Microbiol. 1997. 34:103–113.
4. Hawkins RE, Lissner CR, Sanford JP. Enterobacter sakazakii bacteremia in an adult. South Med J. 1991. 84:793–795.
5. Ongrádi J. Vaginal infection by Enterobacter sakazakii. Sex Transm Infect. 2002. 78:467.
6. Centers for Disease Control and Prevention (CDC). Enterobacter sakazakii infections associated with the use of powdered infant formula-Tennessee, 2001. MMWR Morb Mortal Wkly Rep. 2002. 51:297–300.
7. Block C, Peleg O, Minster N, Bar-Oz B, Simhon A, Arad I, et al. Cluster of neonatal infections in Jerusalem due to unusual biochemical variant of Enterobacter sakazakii. Eur J Clin Microbiol Infect Dis. 2002. 21:613–616.

8. Biering G, Karlsson S, Clark NC, Jónsdóttir KE, Lúdvígsson P, Steingrímsson O. Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J Clin Microbiol. 1989. 27:2054–2056.

9. Drudy D, Mullane NR, Quinn T, Wall PG, Fanning S. Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin Infect Dis. 2006. 42:996–1002.
10. Iversen C, Forsythe S. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci Technol. 2003. 14:443–454.

11. Leclercq A, Wanegue C, Baylac P. Comparison of fecal coliform agar and violet red bile lactose agar for fecal coliform enumeration in foods. Appl Environ Microbiol. 2002. 68:1631–1638.

12. Skladal P, Mascini M, Salvadori C, Zannoni G. Detection of bacterial contamination in sterile UHT milk using an L-lactate biosensor. Enzyme Microb Technol. 1993. 15:508–512.

13. Simmons BP, Gelfand MS, Haas M, Metts L, Ferguson J. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect Control Hosp Epidemiol. 1989. 10:398–401.

14. Questions and answers on Enterobacter sakazakii in powdered infant formula. Version 4. 2004 Feb 13. World Health Organization;
Available from: http://www.who.int/foodsafety/publications/micro/en/qa2.pdf.
15. Yoo MK, Kim SS, Oh SS. Isolation and genotyping of Enterobacter sakazakii from powdered infant formula manufactured in Korea. Food Sci Biotechnol. 2005. 14:875–877.
16. van Acker J, de Smet F, Muyldermans G, Bougatef A, Naessens A, Lauwers S. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J Clin Microbiol. 2001. 39:293–297.

17. Lehner A, Nitzsche S, Breeuwer P, Diep B, Thelen K, Stephan R. Comparison of two chromogenic media and evaluation of two molecular based identification systems for Enterobacter sakazakii detection. BMC Microbiol. 2006. 6:15.
18. Stock I, Wiedemann B. Natural antibiotic susceptibility of Enterobacter amnigenus, Enterobacter cancerogenus, Enterobacter gergoviae and Enterobacter sakazakii strains. Clin Microbiol Infect. 2002. 8:564–578.

19. Lehner A, Tasara T, Stephan R. 16S rRNA gene based analysis of Enterobacter sakazakii strains from different sources and development of a PCR assay for identification. BMC Microbiol. 2004. 4:43.
20. Muytjens HL, van der Ros-van de Repe J. Comparative in vitro susceptibilities of eight Enterobacter species, with special reference to Enterobacter sakazakii. Antimicrob Agents Chemother. 1986. 29:367–370.

21. Iversen C, Waddington M, On SL, Forsythe S. Identification and phylogeny of Enterobacter sakazakii relative to Enterobacter and Citrobacter species. J Clin Microbiol. 2004. 42:5368–5370.

22. Seo KH, Brackett RE. Rapid, specific detection of Enterobacter sakazakii in infant formula using a real-time PCR assay. J Food Prot. 2005. 68:59–63.

23. Malorny B, Wagner M. Detection of Enterobacter sakazakii strains by real-time PCR. J Food Prot. 2005. 68:1623–1627.
24. Kolbert CP, Persing DH. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr Opin Microbiol. 1999. 2:299–305.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download