Abstract
Orofacial clefts, including cleft lip with or without palate (CL/P) and cleft palate (CP), are one of the most common congenital malformations in Asian populations, where the rate of incidence is higher than in European or other racial groups. A number of candidate genes have been identified for orofacial clefts, although no single candidate has been consistently identified in all studies. We performed case-parent trio and case-control studies on 6 single nucleotide polymorphisms (SNPs) in the MSX1 gene using a sample of 52 CL/P and CP probands from Korea. In the case-control study, the allele frequencies of 6 MSX1 SNPs were compared between 52 oral cleft cases and 96 unmatched controls. For the case-parent trio study, single-marker and haplotype-based tests of transmission disequilibrium using allelic and genotypic tests revealed significant evidence of linkage in the presence of disequilibrium for 1170 G/A of exon 2. With the GG genotype as a reference group among GG, GA, and AA genotypes at 1170G/A, the disease risk decreased with the presence of the A allele (AA genotype: OR = 0.26, 95% CI = 0.10-0.99). These results are consistent with evidence from other studies in the US and Chile and confirm the importance of the MSX1 genotype in determining the risk of CL/P and CP in Koreans.
Oral clefts, including cleft lip with or without palate (CL/P) and cleft palate (CP), constitute one of the most common groups of birth defects, affecting up to 1 in 1000 live-born infants worldwide.1 Birth prevalence of CP and CL/P is generally higher in Asian populations compared to European populations. There are 1.81 oral cleft cases per 1000 live births each year in Korea.2 Oral clefts have a complex and heterogeneous etiology; complex in that both genetic and environmental factors control the risk of oral clefts and heterogeneous in that different causal agents are likely present.3,4 Oral clefts show strong familial aggregation and a higher concordance rate among monozygotic twins compared to dizygotic twins, confirming that genetic factors play an important role in the etiology.3 Furthermore, many Mendelian malformation syndromes include oral clefts as a common phenotypic feature.
Several studies have reported an association between the MSX1 gene and oral clefts (CL/P,CP), primarily using an intronic CA-repeat polymorphism.5-10 Knock-out mouse models show CP results from the complete loss of the MSX1 gene product and some linkage studies have provided evidence that the region of chromosome 4 containing MSX1 may contain a causal mutation. Several studies show that polymorphic markers in or near the MSX1 gene on chromosome 4 present as risk factors for clefting, CL/P or CP.9,10 However, no single variant in this gene has been directly implicated as being a causal mutation. In this study, we sequenced the MSX1 gene in a group of Korean controls, identified 6 single nucleotide polymorphisms (SNPs), genotyped the SNPs in 52 case-parents trios and 96 unrelated controls, and then performed family-based and case-control analyses to test for an association between MSX1 and oral clefts.
Fifty-two unrelated Korean patients aged 6 months to 19 years were recruited through the Department of Plastic surgery, Yonsei University Medical center (Seoul, Korea) from January of 2003 to March of 2004. Cases were selected randomly. Enrollees of this study were assigned to the following groups: case only, case with one parent, or triad of case and both parents. Parents of the cases were interviewed regarding family history, medical history, and exposure to suspected risk factors. The patient and his/her medical records were examined to confirm classification as non-syndromic cleft lip with or without palate (CL/P) or cleft palate (CP). The 52 probands included 14 patients with CL, 8 patients with CP, and 30 patients with CLP (cleft lip with palate). A control group was comprised of 96 unrelated Korean adults (48 males and 48 females, ages 23-60) who attended Yonsei University Medical center for a routine medical checkup and did not have an oral cleft or other major health problem. The institutional review boards of Yonsei University and the Johns Hopkins Bloomberg School of Public Health approved this study.
A 9.8-kb region in and around the MSX1 gene was sequenced, including the promoter region but not the intron between exon 1 and exon 2. About 5 kb of the 3' UTR and 5' UTR region of MSX1 were selected, following homology comparison using GenomeVista (http://pipeline.lbl.gov/cgi-bin/GenomeVista).
To identify SNPs present in Koreans, we sequenced the 48 females from the 96 unrelated Korean adults used in the case-control study. Genomic DNA was extracted from leukocytes isolated from whole blood (collected in EDTA) and used as a template for amplification by Polymerase Chain Reaction (PCR). Sequencing was done on the ABI Prism 3700 Genetic Analyzer. Fourteen SNPs were identified in the MSX1 gene by direct sequencing. Forty-eight normal male controls were confirmed with female controls by additional genotyping.
Each SNP was genotyped using the single-base pair primer extension assay with the SNaP shot Multiplex kit (Applied Biosystems, Foster City, CA, USA) as previously reported.11 The results were analyzed using GeneScan analysis, version 3.7, software package (Applied Biosystems, Foster City, CA, USA).
Tests for Hardy-Weinberg equilibrium and case-control comparisons were done using SAS Genetics 9.12.12 A p-value of less than 0.05 was considered statistically significant. Pairwise linkage disequilibrium (LD) was measured using the standardized D' metric.13 Empirical p-values to test for departure from two-loci-linkage equilibrium between alleles at each locus were obtained via permutation tests.
Genotypic TDT analyses14,15 were performed for each marker separately. This approach compares each case's genotype to the other three possible genotypes given his/her parental mating type in a 3:1 matched analysis, using a conditional logistic regression model. This provides a flexible model that includes indicator variables for both heterozygotes and homozygotes separately as:
where X1 and X2 are indicator variables for heterozygotes and homozygotes, respectively. This conditional logistic model was fit using PROC PHREG in SAS (version 9.12), and we used a likelihood ratio test with 2 degrees of freedom to test the null hypothesis of no genotypic effect on the log odds of being a case (Ho: β1 = 0, β2 = 0).
We performed haplotype-based TDT analyses using the HBAT command in the FBAT program.16 This approach estimates haplotype probabilities for individuals with ambiguous phase in families using an EM algorithm, and then tests for strict Mendelian transmission of the haplotype. Rejection of the composite null hypothesis of no association (no LD) or no linkage between the observed phenotypes and the haplotype is evidence of linkage and association.
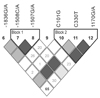
The distribution of cleft types and general characteristics for the cases in this study are given in Table 1. Novel SNPs were confirmed by re-sequencing. To determine genotyping error, haplotype estimates with all triads were performed. Fourteen SNPs were identified by sequencing, including 9 not previously reported in any other study. Of the 14 SNPs, we excluded 3 because of sequencing problems and 5 because the minor allele frequency was under 5% (SNP1 had an allele frequency under 5% but was included because it is in the coding region of MSX1) (Table 2). In total, 6 SNPs were included in this study, which was sufficiently informative (-1836G/A, -1508C/A, -1507G/A, C101G, C330T, and 1170G/A). The locations of these 6 SNPs are shown in Fig. 1. Three of the 6 SNPs also had reasonably high levels of heterozygosity, while 2 SNPs had heterozygosity < 10 %. Pairwise LD measures (D') across all 6 markers of this gene suggest two distinct haplotype blocks in this region, as shown in Fig. 2.
Single-marker TDT analyses were performed on all 6 markers separately. Estimated odds ratios (OR) and respective p-values are shown in Table 3 for the case-parent trio study. This analysis suggested that cases are more likely to be homozygous for the G allele at SNP7. In the genotypic comparison, the GG genotype was significantly associated with disease and the AA genotype with the reference group (OR = 4.1, 95% CI = 1.0-16.8). There was also evidence of a significant trend in risk of G allele (p-value for trend: 0.016). A subset analysis for only CL/P and CP showed a similar association with these SNPs (data not shown).
To test for excessive transmission of certain haplotypes from a parent to a cleft proband, we evaluated haplotypes consisting of 2-4 SNPs using the FBAT program. Taking consecutive markers resulted in 6 windows across the gene (Table 4). The haplotype configuration showing the greatest discrepancy from that expected under the null hypothesis (independent transmission) is shown for each window in Table 4. All haplotype windows containing SNP7 showed significant excessive transmission of a haplotype containing the G allele, although some p-values were only marginally significant (e.g. window 4). The CL/P, CP subset showed a similar but not statistically significant trend of excessive transmission of the haplotypes containing SNP7 (data not shown).
The distribution of the allele frequencies at the 6 markers was different between cases and controls (Table 5). Comparing these 6 markers individually between cases and controls showed a significant difference only for SNP7 (1170G/A). With the GG genotype as a reference group, the disease risk decreased with the presence of the A allele (AA genotype: OR = 0.26, 95% CI = 0.10-0.99). When the A allele was used as the reference group, the risk of the disease increased with the number of G alleles (G allele: OR = 1.97, 95% CI = 1.13-3.44).
We evaluated 6 SNP markers in the MSX1 gene for 52 cleft case-parent trios and a case-control group to assess the association between MSX1 variation and the risk of oral cleft. Oral cleft is one of the most common congenital malformations. The birth prevalence of oral clefts is ~1/800 live births in Korea, which is generally higher than that in European populations or other geographic groups. Evidence from both animal and human studies has shown that MSX1 is involved in the etiology of oral clefts. Although the exact action of MSX1 remains uncertain, it appears to promote growth and inhibit differentiation.17
This study is the first report on the association of MSX1 SNPs and oral clefts in the Korean population. MSX1 has been studied in other ethnic groups and it has been suggested that only a small percentage of clefts are attributed to mutations or variants in this gene.18-21
In the present study, a total of 14 SNPs were identified near or in MSX1, 6 of which were sufficiently polymorphic to be analyzed. Our sequencing concentrated on regions of MSX1 that are thought to be functionally important. Thus, the coding regions and a large conserved mouse sequence homology element received priority. We used the program 'GenomeVista' to find homologous regions between human and mouse. We added more than 4 kb of 5'UTR and 3'UTR to our sequencing efforts to identify additional conserved regulatory regions before running this program.6
Only 1 SNP, 1170G/A in exon 2 (3' UTR), was significantly associated with all three types of clefts in this small Korean population. Allelic association was detected for allele G, which showed a significant trend in risk among all clefts [OR = 1.97, 95% CI = 1.13-3.44; trend p-value: 0.016]. The GG genotype was significantly associated with cleft [OR = 4.10, 95% CI = 1.00-16.80] compared to the AA homozygote. The SNP 1170G/A is not located far from the CA repeat polymorphism previously reported.5,15,22 Haplotypes defined by several SNPS on the same chromosome may provide greater statistical power than single markers for detecting LD with an unobserved high risk allele because they can identify the unique background haplotype carrying the causal mutation. Results from our haplotype analysis confirm the findings of Fallin et al. (2003) and further argue that MSX1 may be a causal gene for oral clefts. Our findings must be interpreted with caution, however. Although MSX1 plays an important role in development, and both animal and human studies have shown that this gene can influence risk for oral clefts, palatogenesis is a complex process that involves many genes.23 Some previous studies showed a significant association between C330T and CL/P, CP especially in Asian populations.6 However, no association between this point mutation and disease was found in the present study. In our case-control study, we concluded that comparing cases with adult controls is appropriate. Non-transmitted parental allele frequencies are similar in both groups with an additional 20 markers (data not shown). The observed association between 1170G/A with CL/P, CP suggests that MSX1 has a role in the developing embryo and this SNP in the 3'UTR region may be crucial for post-transcriptional regulation or mRNA stability. At present, the functional consequences of the MSX1 SNPs are unknown and should be investigated further. The relatively small sample size of our case-control study could lead to false positives and a problem with statistical power. Therefore, our results should be confirmed with a larger sample size. We recruited 96 unrelated controls, but these controls are not representative of the Korean population. We compared the cases' parents to the controls in terms of smoking and drinking and assumed that these controls were comparable, but there is a possibility that our controls may not be a true representation of normal Korean subjects. Despite these complications, one strength of our study is the finding that the genetic structure of Koreans is relatively homogeneous. In further studies, we suggest examining linkage disequilibrium between the previously identified intronic CA repeat and 1170G/A, in order to contribute to our understanding of how MSX1 function affects oral cleft risk. In conclusion, genetic polymorphisms in and around the MSX1 gene showed significant evidence for association from both case-parent trio and case-control anaylsis with Korean samples. The findings contribute to the growing body of evidence that MSX1 plays a role in the etiology of oral clefts. Our results argue that the G allele of 1170G/A in the 3'UTR of MSX1 shows significant evidence for being near the causal mutation and future functional studies should perhaps focus on this region.
Figures and Tables
Fig. 2
Linkage disequilibrium (D' top triangle; all p values < 0.01) between all markers in the MSX1 gene among Korean oral cleft case-parent trios.
References
1. Mossey PA, Little J. Wyszynski DF, editor. Epidemiology of oral clefts: an international perspective. Cleft Lip and Palate: From Origin to Treatment. 2002. New York: Oxford University Press.
2. Kim S, Kim WJ, Oh C, Kim JC. Cleft lip and palate incidence among the live births in the Republic of Korea. J Korean Med Sci. 2002. 17:49–52.
3. Mitchell LE, Beaty TH, Lidral AC, Munger RG, Murray JC, Saal HM, et al. Guidelines for the design and analysis of studies on nonsyndromic cleft lip and cleft palate in humans: summary report from a Workshop of the International Consortium for Oral Clefts Genetics. Cleft Palate Craniofac J. 2002. 39:93–100.
4. Prescott NJ, Winter RM, Malcolm S. Nonsyndromic cleft lip and palate: complex genetics and environmental effects. Ann Hum Genet. 2001. 65:505–515.
5. Fallin MD, Hetmanski JB, Park J, Scott AF, Ingersoll R, Fuernkranz HA, et al. Family-based analysis of MSX1 haplotypes for association with oral clefts. Genet Epidemiol. 2003. 25:168–175.
6. Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet. 2003. 40:399–407.
7. Beaty TH, Wang H, Hetmanski JB, Fan YT, Zeiger JS, Liang KY, et al. A case-control study of nonsyndromic oral clefts in Maryland. Ann Epidemiol. 2001. 11:434–442.
8. Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, et al. Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families. Am J Hum Genet. 2002. 71:349–364.
9. van den Boogaart MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000. 24:342–343.
10. Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, et al. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998. 63:557–568.
11. Turner D, Choudhury F, Reynard M, Railton D, Navarrete C. Typing of multiple single nucleotide polymorphisms in cytokine and receptor genes using SNaP shot. Hum Immunol. 2002. 63:508–513.
12. SAS Institute. SAS release 9.1.2. 2004. Cary, NC, USA: SAS Inc..
13. Lewontin RC. The detection of linkage disequilibrium in molecular sequence data. Genetics. 1995. 140:377–388.
14. Schaid DJ. Likelihoods and TDT for the case-parents design. Genet Epidemiol. 1999. 16:250–260.
15. Beaty TH, Hetmanski JB, Zeiger JS, Fan YT, Liang KY, VanderKolk CA, et al. Testing candidate genes for non-syndromic oral clefts using a case-parent trio design. Genet Epidemiol. 2002. 22:1–11.
16. Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet. 2001. 9:301–306.
17. Odelberg SJ, Kolhoff A, Keating MT. Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000. 103:1099–1109.
18. Hecht JT, Mulliken JB, Blanton SH. Evidence for a cleft palate only locus on chromosome 4 near MSX1. Am J Med Genet. 2002. 110:406–407.
19. Lidral AC, Murray JC, Buetow KH, Basart AM, Schearer H, Shiang R, et al. Studies of the candidate genes TGFB2, MSX1, TGFA and TGFB3 in the etiology of cleft lip and palate in the Philippines. Cleft Palate Craniofac J. 1997. 34:1–6.
20. Stadler HS, Padanilam BJ, Buetow K, Murray JC, Solursh M. Identification and genetic mapping of a homeobox gene to the 4p16.1 region of human chromosome 4. Proc Natl Acad Sci U S A. 1992. 89:11579–11583.
21. Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996. 13:417–421.
22. Vieira AR, Orioli IM, Castilla EE, Cooper ME, Marazita ML, Murray JC. MSX1 and TGFB3 contribute to clefting in South America. J Dent Res. 2003. 82:289–292.
23. Mitchell LE, Murray JC, O'Brien S, Christensen K. Evaluation of two putative susceptibility loci for oral clefts in the Danish population. Am J Epidemiol. 2001. 153:1007–1015.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download