Abstract
Hepatocytes are the primary targets of the hepatitis C virus (HCV). While immunosuppressive roles of HCV core protein have been found in several studies, it remains uncertain whether core protein expressed in hepatocytes rather than in immune cells affects the CD8+ T cell response. In order to transduce genes selectively into hepatocytes, we developed a baculoviral vector system that enabled primary hepatocytes to express a target epitope for CD8+ T cells, derived from ovalbumin (OVA), with or without HCV core protein. Culture of OVA-specific CD8+ T cells with hepatocytes infected with these baculoviral vectors revealed that core protein has no effect on proliferation or apoptosis of CD8+ T cells. Our results suggest that HCV core protein does not exert its suppressive role on the CD8+ T cell immune response through expression in hepatocytes.
The hepatitis C virus causes chronic hepatitis in more than 50% of infected patients.1 One of the mechanisms proposed to explain chronic HCV infection is a weak immune response to HCV, especially the response exerted by CD8+ cytotoxic T lymphocytes (CTL), as the CTL response in HCV infection was found to be weaker and shorter lived in chronically-infected human patients than in individuals who resolved the HCV infection.2,3 Moreover, in the chimpanzee, the only animal model for HCV infection, the animals with strong CTL responses were most likely to resolve the infection.4
Not surprisingly, the mechanisms through which HCV may evade the immune response are of great interest. To date, studies have shown the virus to be highly mutable and capable of generating frequent escape mutations.5,6 HCV proteins may even exert a direct immunosuppressive effect, although data on this point are contradictory. HCV core protein was reported to suppress the CD8+ T cell response in a mouse model using a vaccinia viral vector.7 Later, the same research group showed that interaction between HCV core protein in the blood and the complement receptor specific for the globular heads of the complement C1q protein, gC1qR, inhibited production of IL-12 by human monocyte/macrophages and proliferation of human T cells.8-10 Finally, circulating HCV core protein inhibited effector cytotoxic T cell differentiation.11
In line with the above results, T cells from transgenic mice that expressed the core gene under control of the CD2 promoter showed decreased production of IFN-γ and IL-2.12 In contrast, induced expression of HCV core protein in the liver using an adenovirus vector did not affect priming of CTL, cytokine production, infiltration of lymphocytes into the liver, or liver injury.13 In support of these negative findings, mice were able to clear infections of recombinant adenoviruses that did or did not express HCV core and envelope proteins.14 Due to the apparent contradictions in these data, it is uncertain whether HCV core protein expressed in the hepatocyte, the major site of HCV replication, can regulate the CTL response.
With the intent of clarifying some of the issues surrounding HCV infection and immune response, we developed a baculoviral vector expressing the Enhanced Green Fluorescent Protein (EGFP) and ovalbumin (OVA) peptide for CD8+ T cell recognition, with or without the HCV core protein. The baculoviral vector was chosen for its efficiency in delivering genes to primary hepatocytes and for its hepatocyte tropism.15,16 Our vector system also has the advantage that the antigenic epitope recognized by CD8+ T cells is distinct from the HCV protein under investigation because the epitope is engineered into the marker of target cell infection, EGFP. Using this vector system and a co-culture model in which primary mouse hepatocytes stimulate OVA peptide-specific CD8+ T cells, we found that HCV core protein expressed in primary hepatocytes did not modulate either the proliferation or apoptosis of antigen-specific CD8+ T cells.
To facilitate detection of infected hepatocytes, we constructed baculoviral vectors expressing EGFP. The fragment of pIRES-EGFP (Clontech) between the NsiI and SspI recognition sites, which contains the EGFP-coding gene sequence under the IRES sequence controlled by the CMV promoter, was inserted between the PstI and StuI sites of pAcSG2 (BD PharMingen). Oligonucleotides coding for ovalbumin amino acids 250 through 264 were ligated to the end of the EGFP coding gene sequence using the BsrG1 and NotI sites. The ligation was confirmed by resequencing. The NsiI/SspI fragment of pIRES-EGFP-OVA was then transferred to pAcSG2. The HCV core gene was amplified using HCV 1a cDNA as template (Dr. C. M. Rice, The Rockefeller University) and sense and anti-sense primers containing the NheI and BamHI sites, respectively; 5'-GCCTAGCTAGCATGAGCACGAATCCTAAACCTC-3'; 5'-CGGGATCCTTAGGCTGAAGCGGGCACA-3'. The PCR product was introduced into the pAcSG2-EGFP-OVA vector downstream of the CMV promoter (Fig. 1A). Successful construction of the vector was verified by sequencing.
Recombinant baculoviruses were produced by cotransfecting Sf9 cells with baculoviral Gold DNA (BD PharMingen) and the baculoviral transfer vectors (pAcSG2-EGFP, pAcSG2-EGFP-OVA, and pAcSG2-Core-EGFP-OVA). Recombinant baculoviruses were amplified in Sf9 cells and concentrated by centrifugation of the culture supernatant at 6000 rpm for 16 h at 4℃. The virus pellet was resuspended in DMEM-F12 media (GibcoBRL). To determine the amount of baculovirus for hepatocyte infection, a serial dilution of baculoviruses was added to CHO cells (2 × 104) and the percentage of infected cells showing green fluorescence analyzed by flow cytometry.
The livers of C57BL/6 (B6) mice were perfused via the portal vein with liver perfusion medium (GibcoBRL) and liver digestion medium (Gibco BRL). The released hepatocytes were washed twice by centrifugation at 50 g in order to deplete endothelial and Kupffer cells. Dead cells were removed by Percoll gradient centrifugation. After washing, 2 × 104 hepatocytes were added to each well of 48-well plates (Falcon) and allowed to adhere overnight before being infected with baculovirus vector. The virus was titrated based on its ability to infect CHO cells, and each well was given ten times the amount required for 50% infection of 2 × 104 CHO cells. To visualize the selective infection of hepatocytes with baculovirus, liver cells were prepared as above except with omission of the low speed centrifugation step, allowed to adhere overnight, infected for 24 h with baculovirus expressing EGFP, and labeled with acetylated low-density lipoprotein conjugated to DiI (DiI AcLDL, Molecular Probes) for the final 16 h. DiI AcLDL labels Kupffer and endothelial cells but not hepatocytes.17,18 The cells were photographed using an Olympus fluorescent tissue culture microscope with digital image capture.
Baculovirus-infected hepatocytes were stained on ice for 1 h using 25-D1. 16 hybridoma cell culture supernatant containing antibodies specific to the molecular complex of H-2Kb and SIINFEKL OVA peptide (Dr. R. Germain, National Institutes of Health).19 After washing, cells were incubated with PE-conjugated anti-mouse Ig sera on ice for 30 min, followed by fixation with 2% paraformaldehyde and visualization with a fluorescence microscope.
Baculovirus-infected hepatocytes (2 × 104 cells/well) were co-cultured with a T cell line originally derived from OT-1 mice (2 × 105 cells/well) that had been left untreated for 8 days before the start of co-culture. TCR of this T cell line is specific to the complex of H-2Kb and SIINFEKL OVA peptide. After 2 days, the culture supernatant was harvested and interferon-γ production measured using an ELISA kit (Endogen).
Total RNA of hepatocytes after 24 h of baculoviral infection was isolated using an RNA isolation kit (BD PharMingen). To remove contaminated vector DNA, the RNA samples were treated with DNase I (1U/µl, Invitrogen) for 15 min at room temperature and then reverse transcribed using oligo-dT primers. With cDNA or RNA as a template and the core primer set, PCR was performed using an MJ Research thermal cycler.
Hepatocytes after 24 h of infection were lysed in RIPA buffer [50-mM TrisCl, pH 7.5, 150-mM NaCl, 1-mM EGTA, 0.5% Nonidet P-40, 0.5% deoxycholate, 1-mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, 0.3-µM aprotinin, and 1-µM leupeptin]. Proteins were separated by 15% SDS-PAGE, transferred to polyvinylidene difluoride membranes, and visualized by Western blot analysis using rabbit sera specific to HCV core protein (Dr. Y. C. Sung, Pohang University of Science and Technology, Korea) and the enhanced chemiluminescent system (Amersham Pharmacia Biotech).
Lymph nodes and spleen cells were isolated from OT-1 transgenic mice20 originally provided by Dr. Francis Carbone (University of Melbourne, Australia). These B6 mice expressed TCR specific to the SIINFEKL OVA peptide bound to H-2Kb, to which the CD45.1 marker had been back-crossed. CD8+ T cells were purified using magnetic beads. Briefly, cells were incubated with beads coupled to anti-mouse IgM (Polysciences, Inc., PA) for 30 min on ice, and then those cells attached to the beads were removed with a magnet. Cells were incubated for 30 min at 4℃ with a mixture of antibodies against mouse CD4 (GK1.5), NK1.1 (HB-191), FcγR (2-4G2), and MHC class II molecule (212.A1). After one washing, cells were resuspended in HBSS containing 5% FBS and magnetic beads conjugated to anti-mouse IgG Ab, anti-rat IgG Ab, and anti-mouse IgM Ab. Non-attached cells were harvested. Flow cytometry indicated that the purity of the CD8+ T cells was between 90 and 98%.
To quantitate apoptosis, purified OT-1 CD8+ T cells (2 × 105 cells/well) were added to hepatocyte cultures, and the terminal deoxynucleotidyl-transferase-mediated dUTP-biotin nick end-labeling (TUNEL) assay was conducted according to the manufacturer's instructions (Roche Applied Science). For the proliferation assay, purified OT-1 T cells (2 × 105 cells/well), labeled with 5,6-carboxy-fluorescein diacetate succinimidyl ester (CFSE; Molecular Probe) by incubation of 1 × 107 cells in PBS containing 1-µM CFSE at 37℃ for 10 min, were added to hepatocyte cultures. After co-culture, OT-1 T cells were harvested in PBS containing 10-mM EDTA, washed, and stained with antibodies raised against CD8 and CD45.1 (BD PharMingen). Flow cytometry analysis was performed using CELLQuest™ (Becton Dickinson).
For easy detection of baculovirus-infected hepatocytes, the eukaryotic gene expression cassette for EGFP was introduced into the baculoviral vector. As shown in Fig. 1B, EGFP was expressed by infected hepatocytes. Preferential infection of hepatocytes with baculovirus was demonstrated by labeling of endothelial cells and Kupffer cells with DiI AcLDL (Fig. 1C). Although liver cells were prepared without the usual depletion of non-parenchymal cells, only a few cells were found to be labeled with DiI AcLDL after 40 h in culture. Cells expressing EGFP were large and double nucleated, as is characteristic of hepatocytes, and did not overlap with DiI AcLDL- labeled cells.
We examined OVA peptide presentation by hepatocytes infected with baculovirus using an antibody specific for the molecular complex of H-2Kb and OVA peptide. This Ab stained the cytoplasmic membranes of hepatocytes expressing the EGFP-OVA peptide fusion protein (EGFP-OVA) but not those expressing EGFP alone (Fig. 1D, lower panel). The molecular complex of H-2Kb and OVA peptide on hepatocytes was also recognized by a T cell line derived from OT-1 mice, leading to interferon-γ production (Table 1). We confirmed expression of HCV core protein using RT-PCR and Western blotting (Fig. 1E and F). The RT-PCR product was approximately 600 bp, and the protein 22 kDa, in line with expectations for HCV core RNA and protein. A control PCR reaction from which the reverse transcription step was omitted gave no product, showing that the RT-PCR product was not amplified from contaminating vector DNA.
To compare proliferation of CD8+ T cells in the cultures expressing EGFP-OVA with and without co-expression of HCV core protein, it was necessary to standardize the frequency of hepatocyte infection. This was done by titrating the recombinant viruses using flow cytometry to detect EGFP expression in CHO cells, which are infectible cells like hepatocytes (data not shown). This method is simple and rapid (requiring only 2 days), as compared to the plaque forming unit assay (7 to 10 days). With our flow cytometry method, we were able to obtain similar levels of hepatocyte infection with baculoviruses expressing EGFP alone, EGFP-OVA, and HCV core-EGFP-OVA (Fig. 2A).
Proliferation of OT-1 CD8+ T cells was visualized by monitoring CFSE dilution after co-culture. Proliferation was not observed at 24 h of co-culture, except in the positive control wells pulsed with OVA peptide (data not shown). On the second day of co-culture, the most divisions were seen in T cells responding to hepatocytes pulsed with OVA (Fig. 2B). Control experiments with hepatocytes lacking expression of OVA peptide (expression of EGFP alone or uninfected) indicated that proliferation of OT-1 T cells was antigen specific: OT-1 T cells proliferated in response to hepatocytes expressing EGFP-OVA, while coexpression of HCV core protein did not result in either more or less frequent divisions than did expression of EGFP-OVA alone (Fig. 2). Experiments with less baculovirus similarly showed no difference between EGFP-OVA and HCV core-EGFP-OVA (data not shown).
In chronic hepatitis C patients, apoptosis of T cells in the peripheral blood and liver has been reported to be higher than in normal volunteers.21,22 The proper interpretation of this finding is unclear, however, because after antigenic stimulation, apoptosis of T cells is increased.23,24 We investigated the apoptosis of CD8+ T cells stimulated with hepatocytes expressing OVA alone or OVA with HCV core protein. Analysis 60 h after co-culture indicated that expression of HCV core protein did not affect T cell apoptosis (Fig. 3). Similarly, experiments with a lower amount of baculovirus showed no effect of HCV core protein on T cell apoptosis (data not shown). Finally, we conducted experiments with previously activated T cells, and, like naive cells, these showed no differences in apoptosis when the vector contained the HCV core or EGFP-OVA alone (data not shown). Likewise, the secretion of interferon-γ was no different when T cells responded to the vector containing HCV core (data not shown).
Our results help to resolve apparent contradictions in the literature concerning the immunosuppressive effects of HCV core protein. We found that HCV core protein expressed in hepatocytes does not affect T cell activation, which agrees with a separate study using an adenovirus vector.13 Different conclusions were drawn, however, from experiments in which expression of HCV core protein, as mediated by vaccinia virus, suppressed a CTL immune response in mice.7 These contradictory findings may be due to differences between the adenoviral and vaccinia viral vectors in terms of hepatocyte tropism and infection of immune cells.
We propose that HCV core protein does not alter T cell function when expressed in hepatocytes, but rather, does so when expressed in leukocytes, including T cells. This position is supported by the observation that T cells expressing HCV core protein produced low amounts of cytokines, including IL-2, and showed increased sensitivity to Fas-mediated apoptosis.12,25,26 The role of HCV core protein in myeloid lineage cells is supported by the finding that human dendritic cells transduced with an adenovirus coding for HCV core and E1 proteins were poor stimulators for proliferation of allogeneic and autologous T cells.27 In addition, the proliferation of T cells and IFN-γ production in mixed lymphocyte reactions with HCV core-expressing macrophages were inhibited.28
Rigorous analysis of T cell biology during HCV infection in vivo is difficult because chimpanzee is the only animal model of HCV infection. In liver and blood samples derived from chronic hepatitis C patients, apoptosis of T cells has been reported to be increased in comparison with normal samples.21,22 Also, mitogen-activated monocytes from chronic hepatitis C patients induced CD4+ and CD8+ T cell apoptosis in a co-culture system.29 These studies appear to support the animal and in vitro experiments suggesting that HCV is immunosuppressive through induction of T cell apoptosis.
CD8+ T cell apoptosis has been proposed to occur preferentially in the liver,30 and, although bone marrow-derived cells seem to be the main players in intrahepatic CD8+ T cell apoptosis, Ag expression in liver parenchymal cells is important for promoting the intrahepatic trapping of CD8+ T cells.31 In culture, hepatocytes promoted activation-induced cell death of T cells in an intercellular adhesion molecule-1 (ICAM-1)-dependent manner.32 ICAM-1 is constitutively expressed in hepatic sinusoids and was over-expressed in hepatitis C-infected liver.33,34 This suggests an alternative model in which antigen presentation in hepatocytes promotes T cell apoptosis and HCV infection of the liver causes increased ICAM-1 expression, thereby promoting T cell apoptosis.
Our data are inconsistent with this latter model. While abundant evidence links HCV core expression to T cell apoptosis, in our experimental system in which HCV core expression was strictly limited to hepatocytes, no effect of HCV core protein on either apoptosis or activation of CD8+ T cells was observed. We conclude that, while HCV core protein might manipulate the T cell response by acting in immune cells, it does not do so in hepatocytes.
Figures and Tables
Fig. 1
Baculovirus-mediated gene expression in hepatocytes. (A) Schematic representation of a baculoviral vector expressing HCV core and EGFP-OVA. (B) Primary mouse hepatocytes were observed after 24 h of infection with baculovirus expressing EGFP. (C) Liver cells infected with baculovirus expressing EGFP were incubated with DiI AcLDL (red) for the final 16 hours. Images from a fluorescence tissue culture microscope were merged using Adobe PhotoShop. (D) Hepatocytes infected with baculovirus expressing EGFP alone or EGFP-OVA were stained with Ab specific to the H-2Kb-OVA peptide complex and PE-conjugated secondary Ab (lower panels). (E) PCR was performed using DNase-treated total RNA or cDNA prepared from hepatocytes infected with baculovirus expressing HCV core protein. (F) Extracts of baculovirus-infected hepatocytes were subjected to SDS-PAGE, and HCV core protein was detected by immunoblotting with a specific rabbit antiserum. HCV, hepatitis C virus; EGFP-OVA, enhanced green fluorescent protein-ovalbumin.
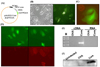
Fig. 2
HCV core protein does not affect proliferation of CD8+ T cells. (A) Comparable levels of infection were demonstrated after 24 h of hepatocyte infection with baculoviruses expressing EGFP alone, EGFP-OVA, and HCV core-EGFP-OVA. (B) CFSE-labeled OT-1 CD8+ T cells were co-cultured with hepatocytes infected with baculovirus or pulsed with OVA peptide. Two days later, T cells were collected and analyzed by FACS. CFSE histograms of OT-1 T cells are gated on CD45.1+ CD8+ cells. Data represent three separate experiments.
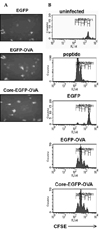
Fig. 3
HCV core protein does not affect apoptosis of CD8+ T cells. OT-1 CD8+ T cells were added to hepatocyte cultures expressing EGFP-OVA with or without HCV core protein. CD8+ T cells were collected, stained, and analyzed by FACS 60 h after co-culture. TUNEL histograms of OT-1 T cells are gated on CD45.1+ CD8+ cells. Data represent three separate experiments.

Table 1
IFN-γ Production by an OT-1 T Cell Line Stimulated with Baculovirus-Infected Hepatocytes

Data represent O.D.
IFN-γ production by an OT-1 T cell line co-cultured with baculovirus-infected hepatocytes was measured by ELISA over 2 days. O.D. of culture with uninfected hepatocytes was 0.127. Similar data were obtained in 6 experiments using OT-1 CD8+ T cells purified from naïve mice; MOI, multiplicity of infection; GFP, green fluorescent protein; OVA, ovalbumin.
ACKNOWLEDGEMENTS
The authors are grateful to Dr. G. Niedermann at the Max-Planck Institute of Immunobiology, Germany, for the kind consultation on our OVA peptide presentation, to Dr. M. F. Mescher at the University of Minnesota for providing us with OT-1 transgenic mice, to Dr. R. N. Germain of the NIH for 25-D1.16 hybridoma cells, to Dr. C. Rice at The Rockefeller University for HCV cDNA, and to Dr. A. Livingstone for the OT-1 T cell line.
References
1. Mattsson L, Sonnerborg A, Weiland O. Outcome of acute symptomatic non-A, non-B hepatitis: a 13-year follow-up study of hepatitis C virus markers. Liver. 1993. 13:274–278.
2. Lechner F, Gruener NH, Urbani S, Uggeri J, Santantonio T, Kammer AR, et al. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol. 2000. 30:2479–2487.
3. Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000. 191:1499–1512.
4. Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY, et al. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999. 10:439–449.
5. Simmonds P. Viral heterogeneity of the hepatitis C virus. J Hepatol. 1999. 31:Suppl 1. 54–60.
6. Yeh BI, Kim HW, Kim HS, Lee JY, Lee KH, Lee KM, et al. The prediction of interferon-alpha therapeutic effect by sequence variation of the HCV hypervariable region 1. Yonsei Med J. 1999. 40:430–438.
7. Large MK, Kittlesen DJ, Hahn YS. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J Immunol. 1999. 162:931–938.
8. Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, Hahn YS. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest. 2000. 106:1239–1249.
9. Eisen-Vandervelde AL, Waggoner SN, Yao ZQ, Cale EM, Hahn CS, Hahn YS. Hepatitis C virus core selectively suppresses interleukin-12 synthesis in human macrophages by interfering with AP-1 activation. J Biol Chem. 2004. 279:43479–43486.
10. Yao ZQ, Eisen-Vandervelde A, Waggoner SN, Cale EM, Hahn YS. Direct binding of hepatitis C virus core to gC1qR on CD4+ and CD8+ T cells leads to impaired activation of Lck and Akt. J Virol. 2004. 78:6409–6419.
11. Accapezzato D, Francavilla V, Rawson P, Cerino A, Cividini A, Mondelli MU, et al. Subversion of effector CD8+ T cell differentiation in acute hepatitis C virus infection: the role of the virus. Eur J Immunol. 2004. 34:438–446.
12. Soguero C, Joo M, Chianese-Bullock KA, Nguyen DT, Tung K, Hahn YS. Hepatitis C virus core protein leads to immune suppression and liver damage in a transgenic murine model. J Virol. 2002. 76:9345–9354.
13. Liu ZX, Nishida H, He JW, Lai MM, Feng N, Dennert G. Hepatitis C virus genotype 1b core protein does not exert immunomodulatory effects on virus-induced cellular immunity. J Virol. 2002. 76:990–997.
14. Sun J, Bodola F, Fan X, Irshad H, Soong L, Lemon SM, et al. Hepatitis C virus core and envelope proteins do not suppress the host's ability to clear a hepatic viral infection. J Virol. 2001. 75:11992–11998.
15. Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci U S A. 1995. 92:10099–10103.
16. Condreay JP, Witherspoon SM, Clay WC, Kost TA. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc Natl Acad Sci USA. 1999. 96:127–132.
17. Nagelkerke JF, Barto KP, van Berkel TJ. In vivo and in vitro uptake and degradation of acetylated low density lipoprotein by rat liver endothelial, Kupffer, and parenchymal cells. J Biol Chem. 1983. 258:12221–12227.
18. Knolle PA, Germann T, Treichel U, Uhrig A, Schmitt E, Hegenbarth S, et al. Endotoxin down-regulates T cell activation by antigen-presenting liver sinusoidal endothelial cells. J Immunol. 1999. 162:1401–1407.
19. Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997. 6:715–726.
20. Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994. 76:17–27.
21. Nuti S, Rosa D, Valiante NM, Saletti G, Caratozzolo M, Dellabona P, et al. Dynamics of intra-hepatic lymphocytes in chronic hepatitis C: enrichment for Vα24+ T cells and rapid elimination of effector cells by apoptosis. Eur J Immunol. 1998. 28:3448–3455.
22. Emi K, Nakamura K, Yuh K, Sugyo S, Shijo H, Kuroki M, et al. Magnitude of activity in chronic hepatitis C is influenced by apoptosis of T cells responsible for hepatitis C virus. J Gastroenterol Hepatol. 1999. 14:1018–1024.
23. Huang L, Soldevila G, Leeker M, Flavell R, Crispe IN. The liver eliminates T cells undergoing antigen-triggered apoptosis in vivo. Immunity. 1994. 1:741–749.
24. Belz GT, Altman JD, Doherty PC. Characteristics of virus-specific CD8(+) T cells in the liver during the control and resolution phases of influenza pneumonia. Proc Natl Acad Sci USA. 1998. 95:13812–13817.
25. Hahn CS, Cho YG, Kang BS, Lester IM, Hahn YS. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology. 2000. 276:127–137.
26. Moorman JP, Prayther D, McVay D, Hahn YS, Hahn CS. The C-terminal region of hepatitis C core protein is required for Fas-ligand independent apoptosis in Jurkat cells by facilitating Fas oligomerization. Virology. 2003. 312:320–329.
27. Sarobe P, Lasarte JJ, Casares N, Lopez-Diaz de Cerio A, Baixeras E, Labarga P, et al. Abnormal priming of CD4(+) T cells by dendritic cells expressing hepatitis C virus core and E1 proteins. J Virol. 2002. 76:5062–5070.
28. Lee CH, Choi YH, Yang SH, Lee CW, Ha SJ, Sung YC. Hepatitis C virus core protein inhibits interleukin 12 and nitric oxide production from activated macrophages. Virology. 2001. 279:271–279.
29. Nakamoto Y, Kaneko S, Kobayashi K. Monocyte-dependent cell death of T lymphocyte subsets in chronic hepatitis C. Immunol Lett. 2001. 78:169–174.
30. Crispe IN, Dao T, Klugewitz K, Mehal WZ, Metz DP. The liver as a site of T-cell apoptosis: graveyard, or killing field? Immunol Rev. 2000. 174:47–62.
31. Mehal WZ, Azzaroli F, Crispe IN. Antigen presentation by liver cells controls intrahepatic T cell trapping, whereas bone marrow-derived cells preferentially promote intrahepatic T cell apoptosis. J Immunol. 2001. 167:667–673.
32. Qian S, Wang Z, Lee Y, Chiang Y, Bonham C, Fung J, et al. Hepatocyte-induced apoptosis of activated T cells, a mechanism of liver transplant tolerance, is related to the expression of ICAM-1 and hepatic lectin. Transplant Proc. 2001. 33:226.
33. Jin Y, Fuller L, Carreno M, Zucker K, Roth D, Esquenazi V, et al. The immune reactivity role of HCV-induced liver infiltrating lymphocytes in hepatocellular damage. J Clin Immunol. 1997. 17:140–153.
34. Horiike N, Onji M, Kumon I, Kanaoka M, Michitaka K, Ohta Y. Intercellular adhesion molecule-1 expression on the hepatocyte membrane of patients with chronic hepatitis B and C. Liver. 1993. 13:10–14.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download