Abstract
The artificial neural network (ANN)–a machine learning technique inspired by the human neuronal synapse system–was introduced in the 1950s. However, the ANN was previously limited in its ability to solve actual problems, due to the vanishing gradient and overfitting problems with training of deep architecture, lack of computing power, and primarily the absence of sufficient data to train the computer system. Interest in this concept has lately resurfaced, due to the availability of big data, enhanced computing power with the current graphics processing units, and novel algorithms to train the deep neural network. Recent studies on this technology suggest its potentially to perform better than humans in some visual and auditory recognition tasks, which may portend its applications in medicine and healthcare, especially in medical imaging, in the foreseeable future. This review article offers perspectives on the history, development, and applications of deep learning technology, particularly regarding its applications in medical imaging.
Machine learning (ML) is defined as a set of methods that automatically detect patterns in data, and then utilize the uncovered patterns to predict future data or enable decision making under uncertain conditions (1). ML is a subset of “artificial intelligence” (AI). In general, there are three approaches to AI: symbolism (rule based, such as IBM Watson), connectionism (network and connection based, such as deep learning or artificial neural net), and Bayesian (based on the Bayesian theorem). The most representative characteristic of ML is that it is driven by data, and the decision process is accomplished with minimum interventions by a human. The program can learn by analyzing training data, and then make a prediction when new data is put in.
Deep learning is a part of ML and a special type of artificial neural network (ANN) that resembles the multilayered human cognition system. Deep learning is currently gaining a lot of attention for its utilization with big healthcare data. Even though ANN was introduced in 1950, there were severe limitations in its application to solve real dilemmas, due to vanishing gradient and overfitting problems, which hindered the training in deep architecture, lack of computing power, and primarily the absence of sufficient data to train the computer system. However, many limitations have now been resolved, given the current availability of big data, enhanced computing power with graphics processing units (GPU), and new algorithms to train a deep neural network (DNN). These deep learning approaches have exhibited impressive performances in mimicking humans in various fields, including medical imaging. One of the typical tasks in radiology practice is detecting structural abnormalities and classifying them into disease categories. Since the 1980s, numerous ML algorithms with different implementations, mathematical bases, and logical theories have been executed to perform such classification tasks. Accordingly, several computer-aided detection (CAD) systems were developed and introduced in the clinical workflow in the early 2000s. However, adverse impacts of these systems have been reported in clinical studies (23). In particular, CAD systems were found to generate more false positives than human readers, which led to a greater assessment time and additional biopsies (2). Thus, the net benefit gained by using CAD was unclear (3). It is expected that current deep learning technology may help overcome the limitations of previous CAD systems, achieve greater detection accuracy, and help make human readers more productive by allowing them to shift humdrum, repetitive radiology tasks to AI.
Deep learning is well suited to medical big data, and can be used to extract useful knowledge from it. This new AI technology has a potential to perform automatic lesion detection, suggest differential diagnoses, and compose preliminary radiology reports. In fact, the globally integrated enterprise IBM is already developing the radiology applications of Dr. Watson. This system includes all the above-mentioned functions, including automatic detection and quantitative feature analysis of the lesion in medical imaging. The rapid rise in AI technology requires radiologists to have knowledge about the technology, in order to understand the ability of AI and how it might change and influence radiologic practice in the near future. We believe that eventually, the adoption of these ML-based analytic tools in radiology practice will happen. However, we also believe that it does not mean a replacement of radiologists, although some specific human tasks will be replaced. These “replacements” will not really be an ultimate replacement, but an overall augmentation of the entire radiology practice, as it will complement irreplaceable and remarkable human skills. In this review, we introduce the history and describe the general, medical, and radiological applications of deep learning.
For training the algorithm, the ML learning methods are classified as supervised learning and unsupervised learning. Supervised learning generates a function that reproduces output by inferring from training data. For this method, training data is prepared with numerical or nominal vectors that represent the characteristics of input data and the corresponding output data. When the output data has a continuous value, the training process is generally referred to as regression. However, if the output data has a categorized value, the process is referred to as classification. In contrast to supervised learning, unsupervised learning does not involve the consideration of output data, but instead infers a function to describe hidden structures from unlabeled input data. Since the examples are unlabeled, there is no objective evaluation of the accuracy. Though unsupervised learning encompasses many other solutions involving summarizing and explaining key features of the data, unsupervised learning is similar to a cluster analysis in statistics, and focuses on the manner which composes the vector space representing the hidden structure, including dimensionality reduction and clustering (Fig. 1).
A naïve Bayesian model that focuses on the probability distribution of input data is a typical classification algorithm. The algorithm is relatively simple, but shows best performance in specific areas such as rRNA sequence assignment (4). The support vector machine (SVM) is the most popular classification algorithm, and typically exhibits the highest performance ranks for most classification problems, given its advantages of regularization and convex optimization (56). Recently, ensemble learning, combined with the diverse classification algorithm for precise prediction, is commonly being used for more advanced classifications (7).
With regard to regression, the linear and logistic regression systems are widely used due to their simple architecture. The parameters of linear regression are estimated to ensure the best fit of the straight line in the data space. Logistic regression employs the logistic function to differentiate binomial distribution, and is usually used as a classifier. Support vector regression (SVR) and ANN are being increasingly used in recent years, and have shown better performances in the regression of certain problems. SVR is a version of SVM for regression (8), and has shown reliable performance in forecasting weather and financial data (910). ANN is a popular regression and classification algorithm for ML, modeling the computational units of multiple layers by imitating signal transmission, and by learning the architecture of neurons and synapses in the human brain.
Figure 2 shows the concept of neural networks derived through biological inspiration. A single neuron consists of dendrites, axon, cell body, and synapse. The simple cell neuron integrates the various input signals and transmits them to other neurons (Fig. 2A). The ANN is composed of interconnected artificial neurons. Each artificial neuron implements a simple classifier model that outputs a decision signal based on the weighted sum of evidences (Fig. 2B). Hundreds of these basic computing units are assembled together to establish the ANN. The weights of the network are trained by a learning algorithm, such as back propagation, where pairs of input signals and desired output decisions are presented, mimicking the condition where the brain relies on external sensory stimuli to learn to achieve specific tasks (11).
Numerical or nominal values used as input data are generally referred to as features in ML. Defining meaningful and powerful features was an important process in previous ML studies. Many domain experts and data scientists sought to discover and generate handcrafted features after applying diverse evaluation approaches, including statistical analysis and performance tests of ML. To enhance this process and achieve training models with higher accuracy, various data cleaning and feature selection methods have been developed to obtain significant improvements in performance. After defining and selecting good handcrafted features, ML algorithms are applied for modeling regression, classification, or unsupervised analysis.
Previous studies show that ANN has remarkable performance in various fields, but had limitations such as a decrease in the local minimum during optimization, and over-training for given values (overfitting). Researchers therefore attempted to use deep architecture to determine solutions, but its complex operation and heavy training costs limited the ability to generate successful models. DNN consists of a series of stacked layers (Fig. 3A). The first layer (input) represents the observed values based on which a prediction is made. The last layer (output) produces a value or class prediction. The layers between the input and output layers are called hidden layers, since their state does not correspond to observable data (input or output). The tiered structure of the neural networks allows them to produce much more complex decisions, based on a combination of simpler decisions. For example, starting with simple localized interpretation of each part of an input, deeper hidden layers can model more complicated networks in the data, thus enabling the classification of a tumor from pixel to curve to shape and to feature. Each edge requires optimized weights for specific training samples. These weights used by DNNs can sum up to billions of parameters, and are randomly initialized and progressively configured by an optimization algorithm such as gradient descent, to find a local minimum of a function by steps proportional to the negative of the gradient of the function at the current point (12). After applying training samples to the network, a loss function between the prediction and the target class or regression value, is quantitatively evaluated. All the parameters are then slightly updated in the direction that will favor the minimization of the loss function.
Based on these neural networks, there are different categories of deep learning with different approaches. DNN extends the depth of layers as compared to traditional ANN, and has shown better performance in prediction and recognition studies, when the layers become complex (13).
Recently, ML researchers have developed technical solutions for implementing deeper architecture (Fig. 3A), as compared to the traditional ANN (Fig. 3B). Using the unsupervised restricted Boltzmann machine (Fig. 4) proposed by Hinton et al. (14), the layers of deep neural architecture were trained separately in an unsupervised manner. As a result, the limitations of DNN, such as local minimum optimization and overfitting, were overcome. As the model could learn from data with deep architecture in an unsupervised manner, it could generate features from raw data. The learning process of this DNN architecture can be observed from the external web based application (15).
The development of hardware technology, such as general-purpose computing on a GPU, has enabled complex operations in shorter computation time for training DNN. Thus, deep learning models now generate meaningful and powerful features after analyzing a large amount of uncategorized data and training the model for accurate prediction by using these features. This process is surprisingly similar to the process of obtaining knowledge in humans with regard to self-organization. These breakthroughs have led to innovative improvements in performances in various research fields, such as speech recognition, image classification, and face recognition. There are several currently available open source deep learning libraries, like Caffe (16), Microsoft Cognitive Toolkit (CNTK) (17), Tensorflow (18), Theano (19), and Torch (20).
The convolutional neural network (CNN), which consists of multiple layers of neuron-like computational connections with step-by-step minimal processing, has achieved significant improvements in the computer vision research area. The overall learning process of CNN simulates the organization of the animal visual cortex (21), and a successfully trained CNN can compose hierarchical information during pre-processing, such as an edge-shape-component-object structure in image classification.
The architecture of CNN is composed of convolutional, pooling layers and fully connected layers (Fig. 5). The primary purpose of a convolutional layer is to detect distinctive local motif-like edges, lines, and other visual elements. The parameters of specialized filter operators, termed as convolutions, are learned. This mathematical operation describes the multiplication of local neighbors of a given pixel by a small array of learned parameters called a kernel (Fig. 6A). By learning meaningful kernels, this operation mimics the extraction of visual features, such as edges and colors, similar to that noted for the visual cortex. This process can be performed by using filter banks. Each filter is a square-shaped object that travels over the given image. The image values on this moving grid are summed using the weights of the filter. The convolutional layer applies multiple filters and generates multiple feature maps. Convolutions are a key component of CNN, and are vital for success in image processing tasks such as segmentation and classification.
To capture an increasingly larger field of view, feature maps are progressively and spatially reduced by pooling the pixels together (Fig. 6B). By propagating only the maximum or average activation through a layer of max or average pooling, convolutional layers subsequently become less sensitive to small shifts or distortions of the target object in extracted feature maps. The pooling layer is used to effectively reduce the dimensions of feature maps, and remain robust to the shape and position of the detected semantic features within the image. In most cases, the max pooling in a feature map is empirically used. These convolutional and pooling layers are repeated several times. The fully connected layers are incorporated to integrate all the feature responses from the entire image and provide the final results. This CNN architecture can be further understood from the external resource (22).
By using deep CNN architecture to mimic the natural neuromorphic multi-layer network, deep learning can automatically and adaptively learn a hierarchical representation of patterns, from low- to high-level features, and subsequently identify the most significant features for a given task (Fig. 5) (23). CNN has the best performance for image classification of a large image repository, such as ImageNet (23).
Because deep CNN architecture generally involves many layers in the neural network, there may be millions of weight parameters to estimate, thus requiring a lot of data samples for model training and parameter tuning. In general, the minimum requirement of data size depends on application of radiologic images. For example, more than 1000 cases per class are needed to train deep learning architecture from scratch in classification. However, there are alternate methods to get around the data size criteria. One is data augmentation, and the other is reuse of the pre-trained network. By using these methods, around 100 cases per class could provide a reasonable outcome.
Recurrent neural network (RNN) is a class of ANN specialized for temporal data including speech and handwriting, where connections between units form a cycle with a one way direction. This creates an internal state of the network which allows it to exhibit dynamic temporal behavior. In contrast to typical neural networks that have structures for a feed-forward network, RNNs can use the temporal memory of networks and yield significant performance improvements in natural language processing, recognition, handwriting recognition, speech recognition and generation tasks (2425).
Use of deep learning has rapidly evolved the field of object recognition in an image. Since the introduction of CNN during the early 2000s, this network has been successfully used for various applications such as traffic sign recognition (26), biological image segmentation (27), and face recognition (28). However, these successes were not well publicized in research and the industry, until the ImageNet open competition in 2012, which contained 1.2 million training images with labels, and 150000 exclusive photographs for validation and testing. The labels represented categories with 1000 distinct objects (29). The challenge involved the development of an efficient ML algorithm to classify images into 1000 object labels. This open competition had an enormous effect and created a new field, wherein researchers compete and collaborate, without having to collect a large-scale labeled dataset. To improve the results of this competition, technical advances such as rectified linear units, a new regularization technique called dropout (30), and a new image augmentation skill (23), were introduced. Moreover, major companies such as Google, Facebook, and Microsoft started to consider deep learning-based image recognition as an important research field. In fact, deep learning techniques achieved a 16% top-5 error rate in 2012 (31), which decreased to below 3% in 2016 (32), and thus surpassed human performance in an object classification task.
The innovations of object classification have been transferred to object localization (33) and semantic segmentation (2834). The CNN-based image recognition framework and RNN-based language model were integrated to establish an image captioning (35), and visual questioning and answering system (36).
Speech recognition is another important area wherein knowledge and research in linguistics, computer science, electrical engineering, and health care, including radiology, can be incorporated. Many researchers (37383940) have developed technologies that enable the recognition and translation of the spoken language into text by computerized devices, including smart technologies and robotics. In recent years, the field of speech recognition has made considerable progress due to advances in deep learning and big data. This is evident by the various papers published in the research field, and the currently available speech recognition systems of many corporations, such as Google, Apple, and Microsoft.
Deep learning techniques have recently been introduced for medical image analysis, with promising results in various applications such as segmentation and registration. Considerable interest has been given to DNNs, particularly CNNs, to resolve the problems associated with medical imaging segmentation. These include approaches for the segmentation of the lungs (41), tumors and other structures in the brain (4243), biological cells and membranes (2744), tibial cartilage (45), bone tissue (46), and cell mitosis (47). All these applications are mostly use two-dimensional (2D) CNN techniques, which take intensity patches as inputs; occasionally, spatial consistency is enforced at a second stage through post-processing computations, such as probabilistic graphical models. However, the time required to train patch-based methods may make the approach infeasible, especially especially with a large size and number of patches.
Recently, different CNN architectures (34484950) have been proposed that feed through entire images; this obviates the need to select representative patches, and eliminates redundant calculations where patches overlap, thus facilitating scale up of such models more efficiently, with better image resolution. Kang and Wang (48) introduced the fully CNN (fCNN) for the segmentation of crowds in surveillance videos. However, fCNNs produce segmentations of lower resolution as compared to input images, due to the successive use of convolutional and pooling layers, both of which reduce the dimensionality. To predict segmentations of the same resolution as the input images, Brosch et al. (5051) recently proposed the use of a 3-layer convolutional encoder network for multiple sclerosis lesion segmentation. The combination of convolutional (52) and deconvolutional (53) layers allows the network to produce segments that are of the same resolution as the input images. This fCNN architecture can also be applied for lesion localization and semantic segmentation. Figure 7 shows the preliminary results of semantic segmentation of knee magnetic resonance (MR) images. In this fCNN based semantic segmentation, a highly accurate lesion probability map can be obtained in fully convolutional layers, even though it is of low resolution. This low resolution map interpolates to achieve the same resolution as that of the input MR image. The weight parameters in this interpolation were further optimized in the training process.
A few studies have assessed the problems associated with medical image registration. In recent years, promising results for object matching in computer vision tasks have been reported via ML methods (54555657). Although these methods can reliably recover the object's location and/or position for computer vision tasks, they are unable to meet the accuracy requirement of 2D/three-dimensional (3D) registration tasks in medical imaging, which often target for very high accuracy (i.e., sub-millimeter) for diagnosis and surgery guidance purposes. Shun Miao et al. (58) proposed a CNN regression approach, referred to as Pose Estimation via Hierarchical Learning (PEHL), to achieve real-time 2D/3D registration, with a large capture range and high accuracy. To capture large and complex deformation in image registration, Zhao and Jia (59) proposed a two-layer-deep adaptive registration framework that first accurately classified the rotation parameter through multilayer CNNs, and then separately identified the scale and translation parameters.
Many advances have also been made in the automatic generation of image captions, to describe contents in an image. Although the applications of previous studies on image caption generation (606162636465666768) were limited to natural image caption datasets, such as Flickr8k (69), Flickr30k (70), or Microsoft Common Objects in Context (MS COCO) (71) in the medical field, continuous effort and progress has been ensured for the automatic recognition and localization of specific diseases and organs, primarily with datasets where the target objects are explicitly annotated (727374757677).
Inspired by early research on image caption generation (787980), studies have recently introduced the use of CNNs and RNNs (606162636465666768) to combine recent advances in computer vision and machine translation, and thus automatically annotate chest radiographs with diseases and descriptions of the context of a disease (e.g., the location, severity, and the affected organs) (81). Thus, authors employ a publicly available radiology dataset of chest radiographs and their reports, and use its image annotations to mine disease names to train the CNNs. To circumvent the large bias between normal vs. diseased, various regularization techniques are adapted to CNNs. RNNs are then trained to describe the context of a detected disease, based on the deep CNN features.
Many different types of CAD systems have been recently implemented as part of picture archiving and communication system (PACS) solutions (82838485). This seamless integration of CAD into PACS increases the reader sensitivity, without significantly increasing image reading time and, thus, improving the efficiency of daily radiology practice.
Briefly speaking, current CAD systems consist of two different parts: detection and false-positivity reduction. Typically speaking, detection is primarily based on algorithms specific to the detection task, resulting in many candidate lesions. The latter part is commonly based on traditional ML to reduce the false positive lesions. Unfortunately, even with these complicated and sophisticated programs, the general performance of current CAD systems is not good, thus hampering their widespread usage in routine clinical practice. Another important limitation of the current CAD systems is susceptibility to the imaging protocols and noise. With its known flexibility to image noise and variation of imaging protocols, deep learning has a potential to improve the performance of current CAD to the level useful in daily practice. In contrast to the current CAD system, deep learning method may provide us a single step solution of CAD. In addition, the unique nature of transfer learning may accelerate the development of the CAD system for various diseases and different modalities.
Early reports of deep learning based CAD systems for breast cancer (86), lung cancer (8788) and Alzheimer's disease (AD) (899091) show promising results regarding their performance in detecting and staging the diseases. Deep learning has been applied for the identification, detection, and diagnosis and risk analysis of breast cancer (869293).
Several deep learning based studies have assessed the implementation of lung cancer screening CAD systems (87888990919293949596979899), and show the potential for predicting lung cancer and classifying lung nodules (9294).
The early detection and diagnosis of AD is also important for patient treatment. Single photon emission computed tomography and positron emission tomography are commonly used by physicians for diagnosis of AD. Few studies have incorporated deep learning based approaches for AD diagnosis and in these systems, wherein the diseases can be assessed from multi-modal brain data due to the effective features generated from deep learning (899091).
Speech recognition applications include voice user interfaces, such as voice dialing, natural language processing, and speech-to-text for radiologic reporting (which has been proven to be a natural interaction modality and effective technology for medical reporting), particularly in the field of radiology. An automatic radiological dictation system was previously used in the radiology field (37). However, at present, the Dragon™(Nuance Communications, Burlington, MA, USA), which is well-known for the development of the Siri voice recognizer, is used for automatic voice dictation and in the reading assistant system in the United States of America and Europe. It is particularly useful for automatic transcription with the devices, and without the need for typing of dictation content from radiologists. SpeechRite™ (Capterra, Arlington, VA, USA) (95) is one of the medical radiologic applications based on the cloud computing architecture that has minimal resource requirements for deployment, and enables remote accessibility by users, thus facilitating the delivery of highly accurate drafts. Moreover, 2Ascribe™ (2Ascribe Inc., Toronto, Canada) (96) offers quality medical transcription for all radiology modalities via speech recognition. The radiologist can train the system and edit their own documents, or the ‘raw’ documents can be edited by qualified medical transcriptionists before being returned. In addition, a specialized microphone, the PowerScribe360® (Nuance Communications) for dictating radiology reports (97), is used for automatic radiological dictation in the medical field. These systems have been used by an increasing number of institutions and physicians with varying degrees of success. In Korea, the development of speech recognition software is more difficult, as the radiologists use a mixture of Korean and English for recording. However, several companies have succeeded in achieving promising initial performance with DNN and RNN.
The original concept of precision medicine involves the prevention and treatment strategies that consider individual variability (98) by assessing large sets of data, including patient information, medical imaging, and genomic sequences. The success of precision medicine is largely dependent on robust quantitative biomarkers. In general, deep learning can be used to explore and create quantitative biomarkers from medical big data obtained through internet of things, genetics and genomics, medicinal imaging, and mobile monitoring sources (Fig. 9). In particular, imaging is non-invasively and routinely performed for clinical practice, and can be used to compute quantitative imaging biomarkers. Many radiomic studies have correlated imaging biomarkers with the genomic expression or clinical outcome (99100). Deep learning techniques can be used to generate more reliable imaging biomarkers for precision medicine.
Even with many promising results from early research studies, there are multiple issues to be resolved before the introduction of deep learning methods in radiological practice, some of which are listed as follows:
Firstly, the high dependency on the quality and amount of training data, and the tendency of overfitting, should be considered. Considering the differences in disease prevalence, imaging machines, and imaging protocols in hospitals all across the worlds, how can we confirm that the developed methods are generally useful? The evaluation methods to test the performance of each technique therefore requires development.
Secondly, the black box nature of the current deep learning technique should be considered. Even when the deep learning based method shows excellent results, in many occasions, it is difficult or mostly impossible to explain the technical and logical bases of the system. Is it acceptable to us to use the system, in this era of ‘evidence based radiology’? Is there any chance for the system failure in rare disease condition?
Thirdly, there could be legal and ethical issues regarding the use of clinical imaging data for the commercial development of deep learning based system, since the performance of the system will be highly dependent on the quality of the data.
Additionally, the legal liability issues would be raised if we were to adopt a deep learning system in certain process of radiological practice, independent from the supervision of a radiologist. As we expect, any system cannot be perfect. Who or what should take the responsibility in case of an error and misinformation that leads to patient harm?
At present, radiologists experience an increasing number of complex imaging tests. This makes it difficult to finish reading in time and provide accurate reports. However, the new technology of deep learning is expected to help radiologists provide a more accurate diagnosis, by delivering quantitative analysis of suspicious lesions, and may also enable a shorter time for reading due to automatic report generation and voice recognition, both of which are benefits that AI can provide in the clinical workflow.
In this review, we introduced deep learning from a radiology perspective. Deep learning has already shown superior performance than humans in some audio recognition and computer vision tasks. This has enabled the development of digital assistants such as Apple's Siri, Amazon's Echo, and Google's Home, along with numerous innovations in computer vision technologies for autonomous driving. Technology giants such as Google, Facebook, Microsoft, and Baidu have begun research on the applications of deep learning in medical imaging. Although it is always difficult to predict the future, these technological changes make it reasonable to think that there might be some major changes in radiology practices in a few decades due to AI. However, when we consider the use of AI in medical imaging, we anticipate this technological innovation to serve as a collaborative medium in decreasing the burden and distraction from many repetitive and humdrum tasks, rather than replacing radiologists.
The use of deep learning and AI in radiology is currently in the stages of infancy. One of the most important factors for the development of AI and its proper clinical adoption in radiology would be a good mutual understanding of the technology, and the most appropriate form of radiology practice and workflow by both radiologists and computer scientists/engineers. With the recent technological innovations by ImageNet, large and fully annotated databases are needed for advancing AI development in medical imaging. This will be vital for training the deep learning network, and also for its evaluation. The active involvement of many radiologists is also essential for establishing a large medical imaging database. Furthermore, there are various other issues and technical problems to solve and overcome. Finally, ethical, regulatory, and legal issues raised in the use of patient clinical image data for the development of AI should be carefully considered. This is another important topic that needs to be discussed among radiologists, scientists/engineers, and law and ethics experts altogether.
Figures and Tables
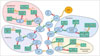 | Fig. 1Categories of machine learning, including classification, regression, clustering, and dimensionality reduction.Adapted from http://scikit-learn.org/stable/tutorial/machine_learning_map/(101). GMM = Gaussian mixture model, LLE = locally-linear embedding, PCA = principal component analysis, SGD = stochastic gradient descent, SVC = support vector classification, SVR = support vector regression, VBGMM = variational Bayesian Gaussian mixture model
|
 | Fig. 3Comparison between shallow learning and deep learning in neural network.
A. Typical deep learning neural network with 3 deep layers between input and output layers. B. Typical artificial neural network with 1 layer between input and output layers.
|
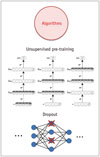 | Fig. 4Two breakthrough algorithms in deep learning, including unsupervised pre-training and dropout. |
 | Fig. 5Architecture of convolutional neural networks, including input, Conv., and FC layers.Conv. = convolutional, FC = fully connected
|
 | Fig. 6Illustration of convolution and pooling methods.
A. Convolution method. B. Max and average pooling methods.
|
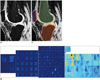 | Fig. 7Example of semantic segmentation in knee MR image.
A. Input MR knee image. B. Feature response maps on layers with different depth in fCNN. C. Output result from fCNN. fCNN = fully convolutional neural network
|
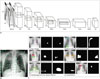 | Fig. 8Preliminary results of lesion detection on chest radiographs, by using faster R-CNN architecture.Each result set is composed of 3 rows. First row shows faster R-CNN results, and ground truth lesion mask is delineated by radiologists in second row. Automatic description is provided in third row.
A. Faster R-SNN architecture. B. Proposed regions of interest. C. Multiple lesion detection results. R-CNN = regional-convolutional neural network
|
References
1. Murphy KP. Machine learning: a probabilistic perspective. 1st ed. Cambridge: The MIT Press;2012. p. 25.
2. Fenton JJ, Taplin SH, Carney PA, Abraham L, Sickles EA, D'Orsi C, et al. Influence of computer-aided detection on performance of screening mammography. N Engl J Med. 2007; 356:1399–1409.
3. Lehman CD, Wellman RD, Buist DS, Kerlikowske K, Tosteson AN, Miglioretti DL, et al. Diagnostic accuracy of digital screening mammography with and without computer-aided detection. JAMA Intern Med. 2015; 175:1828–1837.
4. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007; 73:5261–5267.
5. Byvatov E, Fechner U, Sadowski J, Schneider G. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J Chem Inf Comput Sci. 2003; 43:1882–1889.
6. In : Tong S, Chang E, editors. Support vector machine active learning for image retrieval. Proceedings of the 9th ACM International Conference on Multimedia; 2001 October 5-September 30; Ottawa, Canada. New York: ACM;2001. p. 107.
7. Arbib MA. The handbook of brain theory and neural networks. 2nd ed. Boston: The MIT Press;2003.
8. Drucker H, Burges CJC, Kaufman L, Smola A, Vapnik V. Support vector regression machines. Adv Neural Inf Process Syst. 1997; 9:155–161.
9. Yu PS, Chen ST, Chang IF. Support vector regression for real-time flood stage forecasting. J Hydrol (Amst). 2006; 328:704–716.
10. Tay FEH, Cao L. Application of support vector machines in financial time series forecasting. Omega (Westport). 2001; 29:309–317.
11. Haykin SS. Neural networks: a comprehensive foundation. New York: Macmillan College Publishing;1994. p. 107–116.
12. Kiwiel KC. Convergence and efficiency of subgradient methods for quasiconvex minimization. Math Program. 2001; 90:1–25.
13. In : Deng L, Hinton G, Kingsbury B, editors. New types of deep neural network learning for speech recognition and related applications: an overview. Proceedings of 2013 IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP); 2013 May 26-31; Vancouver, Canada. IEEE;2013. p. 8599–8603.
14. Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006; 18:1527–1554.
15. Playground.tensorflow.org Web site. A Neural Network Playground-TensorFlow. Accessed April 1, 2017. http://playground.tensorflow.org.
16. In : Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, editors. Caffe: convolutional architecture for fast feature embedding. Proceedings of the 22nd ACM International Conference on Multimedia; 2014 November 3-7; Orlando, FL, USA. New York: ACM;2014. p. 675–678.
17. Yu D, Eversole A, Seltzer ML, Yao K, Huang Z, Guenter B, et al. An introduction to computational networks and the computational network toolkit. New York: Microsoft Research;2014.
18. In : Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, editors. TensorFlow: a system for large-scale machine learning. Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation; 2016 November 2-4; Savannah, GA, USA. Berkeley: USENIX Association;2016. p. 265–283.
19. The Theano Development Team. Al-Rfou R, Alain G, Almahairi A, Angermueller C, Bahdanau D, et al. Theano: a python framework for fast computation of mathematical expressions. ArXiv org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1605.02688.
20. In : Collobert R, van der Maaten L, Joulin A, editors. Torchnet: an open-source platform for (deep) learning research. Proceedings of the 33rd International Conference on Machine Learning (ICML-2016); 2016 June 19-24; New York, NY, USA. New York: JMLR;2016.
21. Hubel DH, Wiesel TN. Receptive fields and functional architecture of monkey striate cortex. J Physiol. 1968; 195:215–243.
22. Ujjwalkarn.me Web site. An intuitive explanation of convolutional neural networks. Accessed April 1, 2017. https://ujjwalkarn.me/2016/08/11/intuitive-explanation-convnets.
23. In : Krizhevsky A, Sutskever I, Hinton GE, editors. ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems; 2012 December 3-6; Lake Tahoe, NV, USA. Curran Associates Inc.;2012. p. 1–9.
24. In : Mikolov T, Karafiát M, Burget L, Cernocký J, Khudanpur S, editors. Recurrent neural network based language model. Proceedings of the 11th Annual Conference of the International Speech Communication Association (INTERSPEECH 2010); 2010 September 26-30; Makuhari, Japan. International Speech Communication Association;2010. p. 1045–1048.
25. In : Gregor K, Danihelka I, Graves A, Rezende DJ, Wierstra D, editors. DRAW: a recurrent neural network for image generation. Proceedings of the 32nd International Conference on Machine Learning (ICML-2015); 2015 July 6-11; Lille, France. JMLR;2015. p. 1462–1471.
26. Cireşan D, Meier U, Masci J, Schmidhuber J. Multi-column deep neural network for traffic sign classification. Neural Netw. 2012; 32:333–338.
27. Ning F, Delhomme D, LeCun Y, Piano F, Bottou L, Barbano PE. Toward automatic phenotyping of developing embryos from videos. IEEE Trans Image Process. 2005; 14:1360–1371.
28. Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Semantic image segmentation with deep convolutional nets and fully connected CRFs. ArXiv.org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1412.7062.
29. Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015; 115:211–252.
30. Dahl GE, Sainath TN, Hinton GE. Improving deep neural networks for LVCSR using rectified linear units and dropout. In : Proceedings of the 38th IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP '13); 2013 May 26-31; Vancouver, Canada. IEEE;2013. p. 8609–8613.
31. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In : Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in neural information processing system 25. Nevada: Curran Associates Inc.;2012. p. 1097–1105.
32. Image-net.org Website. Large Scale Visual Recognition Challenge 2016 (ILSVRC2016) results. Accessed April 1, 2017. http://image-net.org/challenges/LSVRC/2016/.
33. In : Ren S, He K, Girshick R, Sun J, editors. Faster R-CNN: towards real-time object detection with region proposal networks. Proceedings of the 28th International Conference on Neural Information Processing Systems (NIPS-2015); 2015 December 7-12; Montreal, Canada. Cambridge: MIT Press;2015. p. 91–99.
34. Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017; 39:640–651.
35. In : Johnson J, Karpathy A, Fei LF, editors. DenseCap: fully convolutional localization networks for dense captioning. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 June 27-30; Seattle, WA, USA. IEEE;2016. p. 4565–4574.
36. In : Antol S, Agrawal A, Lu J, Mitchell M, Batra D, Zitnick CL, editors. VQA: visual question answering. Proceedings of the IEEE International Conference on Computer Vision (ICCV); 2015 December 7-13; Santiago, Chile. IEEE;2015. p. 2425–2433.
37. Antoniol G, Fiutem R, Flor R, Lazzari G. Radiological reporting based on voice recognition. In : In : Bass LJ, Gornostaev J, Unger C, editors. Human-Computer Interaction. Proceedings of the third International Conference, EWHCI '93 Moscow; 1993 August 3–7; Moscow, Russia. Moscow: Springer;1993. p. 242–253.
38. Antoniol G, Brugnara F, Dalla Palma F, Lazzari G, Moser E. A.RE.S.: an interface for automatic reporting by speech. In : Adlassnig KP, Grabner G, Bengtsson S, Hansen R, editors. Proceedings of Medical Informatics Europe 1991. 1991 August 19-22; Vienna, Austria. Berlin, Heidelberg: Springer-Verlag;1991. p. 150–154.
39. Bahl LR, Jelinek F, Mercer RL. A maximum likelihood approach to continuous speech recognition. IEEE Trans Pattern Anal Mach Intell. 1983; 5:179–190.
40. Baker JK. Trainable grammars for speech recognition. J Acoust Soc Am. 1979; 65:S1. S132.
41. Middleton I, Damper RI. Segmentation of magnetic resonance images using a combination of neural networks and active contour models. Med Eng Phys. 2004; 26:71–86.
42. Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016; 35:1240–1251.
43. Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJ, Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016; 35:1252–1261.
44. In : Ciresan D, Giusti A, Gambardella LM, Schmidhuber J, editors. Deep neural networks segment neuronal membranes in electron microscopy images. Proceedings of the 25th International Conference on Neural Information Processing Systems; 2012 December 3-6; Lake Tahoe, NV, USA. Curran Associates Inc.;2012. p. 2843–2851.
45. Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv. 2013; 16(Pt 2):246–253.
46. Glavan CC, Holban S. Segmentation of bone structure in X-ray images using convolutional neural network. Adv Electr Comput Eng. 2013; 13:87–94.
47. Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv. 2013; 16(Pt 2):411–418.
48. Kang K, Wang X. Fully convolutional neural networks for crowd segmentation. ArXiv.org Web site. Accessed April 1, 2017. http://arxiv.org/abs/1411.4464.
49. Roura E, Oliver A, Cabezas M, Valverde S, Pareto D, Vilanova JC, et al. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology. 2015; 57:1031–1043.
50. Brosch T, Yoo Y, Tang LYW, Li DKB, Traboulsee A, Tam R. Deep convolutional encoder networks for multiple sclerosis lesion segmentation. In : Navab N, Hornegger J, Wells WM, Frangi AF, editors. Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015. New York. Spinger;2015. p. 3–11.
51. Brosch T, Tang LY, Yoo Y, Li DK, Traboulsee A, Tam R. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging. 2016; 35:1229–1239.
52. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998; 86:2278–2324.
53. In : Zeiler MD, Taylor GW, Fergus R, editors. Adaptive deconvolutional networks for mid and high level feature learning. Proceedings of the 2011 International Conference on Computer Vision; 2011 November 6-13; Barcelona, Spain. Washington, DC: IEEE Computer Society;2011. p. 2018–2025.
54. In : Wohlhart P, Lepetit V, editors. Learning descriptors for object recognition and 3D pose estimation. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 3109–3118.
55. In : Dollár P, Welinder P, Perona P, editors. Cascaded pose regression. Proceedings of 2010 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2010 June 13-18; San Francisco, CA, USA. IEEE;2010. p. 1078–1085.
56. In : Zach C, Sanchez AP, Pham MT, editors. A dynamic programming approach for fast and robust object pose recognition from range images. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 196–203.
57. In : Mottaghi R, Xiang Y, Savarese S, editors. A coarse-to-fine model for 3D pose estimation and sub-category recognition. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 418–426.
58. Shun Miao, Wang ZJ, Rui Liao. A CNN regression approach for real-time 2D/3D registration. IEEE Trans Med Imaging. 2016; 35:1352–1363.
59. Zhao L, Jia K. Deep Adaptive Log-demons: diffeomorphic image registration with very large deformations. Comput Math Methods Med. 2015; 2015:836202.
60. Mao J, Xu W, Yang Y, Wang J, Huang Z, Yuille A. Explain images with multimodal recurrent neural networks. ArXiv.org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1410.1090.
61. Socher R, Karpathy A, Le QV, Manning CD, Ng AY. Grounded compositional semantics for finding and describing images with sentences. Trans Assoc Comput Linguist. 2014; 2:207–218.
62. Kiros R, Salakhutdinov R, Zemel RS. Unifying visual-semantic embeddings with multimodal neural language models. ArXiv. org Web site. Accessed April 1, 2017. https://arxiv.org/abs/1411.2539.
63. Donahue J, Hendricks LA, Rohrbach M, Venugopalan S, Guadarrama S, Saenko K, et al. Long-term recurrent convolutional networks for visual recognition and description. IEEE Trans Pattern Anal Mach Intell. 2017; 39:677–691.
64. In : Vinyals O, Toshev A, Bengio S, Erhan D, editors. Show and tell: a neural image caption generator. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 3156–3164.
65. In : Fang H, Gupta S, Iandola F, Srivastava R, Deng L, Dollár P, editors. From captions to visual concepts and back. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 1473–1482.
66. Chen X, Zitnick CL. Mind's eye: a recurrent visual representation for image caption generation. In : Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 2422–2431.
67. Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994; 5:157–166.
68. Karpathy A, Li FF. Deep visual-semantic alignments for generating image descriptions. IEEE Trans Pattern Anal Mach Intell. 2016; 39:664–676.
69. Hodosh M, Young P, Hockenmaier J. Framing image description as a ranking task: data, models and evaluation metrics. J Artif Intell Res. 2013; 47:853–899.
70. Young P, Lai A, Hodosh M, Hockenmaier J. From image descriptions to visual denotations: new similarity metrics for semantic inference over event descriptions. Trans Assoc Comput Linguist. 2014; 2:67–78.
71. Lin TY, Maire M, Belongie S, Hays J, Perona P, Ramanan D, et al. Microsoft COCO: common objects in context. In : Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. Computer Vision-ECCV 2014. New York. Springer;2014. p. 740–755.
72. Shi Y, Suk HI, Gao Y, Shen D. Joint coupled-feature representation and coupled boosting for AD diagnosis. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2014; 2014:2721–2728.
73. In : Hofmanninger J, Langs G, editors. Mapping visual features to semantic profiles for retrieval in medical imaging. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 457–465.
74. In : Subbanna N, Precup D, Arbel T, editors. Iterative multilevel MRF leveraging context and voxel information for brain tumour segmentation in MRI. Proceedings of the 27th IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2014); 2014 June 23-28; Columbus, OH, USA. IEEE;2014. p. 400–405.
75. In : Ngo TA, Carneiro G, editors. Fully automated non-rigid segmentation with distance regularized level set evolution initialized and constrained by deep-structured inference. Proceedings of 2014 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2014 June 23-28; Columbus, OH, USA. IEEE;2014. p. 3118–3125.
76. In : Ledig C, Shi W, Bai W, Rueckert D, editors. Patch-based evaluation of image segmentation. Proceedings of 2014 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2014 June 23-28; Columbus, OH, USA. IEEE;2014. p. 3065–3072.
77. In : Rupprecht C, Peter L, Navab N, editors. Image segmentation in twenty questions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 June 7-12; Boston, MA, USA. IEEE;2015. p. 3314–3322.
78. Kulkarni G, Premraj V, Ordonez V, Dhar S, Li S, Choi Y, et al. Babytalk: understanding and generating simple image descriptions. IEEE Trans Pattern Anal Mach Intell. 2013; 35:2891–2903.
79. In : Feng Y, Lapata M, editors. How many words is a picture worth? Automatic caption generation for news images. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics; 2010 July 11-16; Uppsala, Sweden. Stroudsburg: Association for Computational Linguistics;2010. p. 1239–1249.
80. Farhadi A, Hejrati M, Sadeghi MA, Young P, Rashtchian C, Hockenmaier J, et al. Every picture tells a story: generating sentences from images. In : Daniilidis K, Maragos P, Paragios N, editors. Computer Vision-ECCV 2010. Berlin, Heidelberg. Springer;2010. p. 15–29.
81. In : Shin HC, Roberts K, Lu L, Fushman DD, Yao J, Summers RM, editors. Learning to read chest X-rays: recurrent neural cascade model for automated image annotation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016 June 27-30; Las Vegas, NV, USA. IEEE;2016.
82. Bogoni L, Ko JP, Alpert J, Anand V, Fantauzzi J, Florin CH, et al. Impact of a computer-aided detection (CAD) system integrated into a picture archiving and communication system (PACS) on reader sensitivity and efficiency for the detection of lung nodules in thoracic CT exams. J Digit Imaging. 2012; 25:771–781.
83. Welter P, Hocken C, Deserno TM, Grouls C, Günther RW. Workflow management of content-based image retrieval for CAD support in PACS environments based on IHE. Int J Comput Assist Radiol Surg. 2010; 5:393–400.
84. Le AH, Liu B, Huang HK. Integration of computer-aided diagnosis/detection (CAD) results in a PACS environment using CAD-PACS toolkit and DICOM SR. Int J Comput Assist Radiol Surg. 2009; 4:317–329.
85. Zhou Z. Data security assurance in CAD-PACS integration. Comput Med Imaging Graph. 2007; 31:353–360.
86. Wang D, Khosla A, Gargeya R, Irshad H, Beck AH. Deep learning for identifying metastatic breast cancer. ArXiv.org Web site. Accessed April 1, 2017. https://arxiv.org/pdf/1606.05718.pdf.
87. Hua KL, Hsu CH, Hidayati SC, Cheng WH, Chen YJ. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther. 2015; 8:2015–2022.
88. In : Kumar D, Wong A, Clausi DA, editors. Lung Nodule Classification Using Deep Features in CT Images. Proceedings of 2015 12th Conference on Computer and Robot Vision; 2015 June 3-5; Halifax, Canada. IEEE;2015. p. 133–138.
89. Suk HI, Lee SW, Shen D. Alzheimer's Disease Neuroimaging Initiative. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage. 2014; 101:569–582.
90. Suk HI, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv. 2013; 16(Pt 2):583–590.
91. In : Liu S, Lis S, Cai W, Pujol S, Kikinis R, Feng D, editors. Early diagnosis of Alzheimer's disease with deep learning. Proceedings of the IEEE 11th International Symposium on Biodmedical Imaging; 2014 April 29-May 2; Beijing, China. IEEE;2014. p. 1015–1018.
92. Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep. 2016; 6:24454.
93. Kallenberg M, Petersen K, Nielsen M, Ng AY, Pengfei Diao, Igel C, et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging. 2016; 35:1322–1331.
94. Chen J, Chen J, Ding HY, Pan QS, Hong WD, Xu G, et al. Use of an artificial neural network to construct a model of predicting deep fungal infection in lung cancer patients. Asian Pac J Cancer Prev. 2015; 16:5095–5099.
95. Capterra.com Web site. Spantel. SpeechRite. Accessed April 1, 2017. http://www.capterra.com/speech-recognition-software/spotlight/142035/SpeechRite/Spantel.
96. 2ascribe.com Web site. Radiology Dictation & Transcription. Accessed April 1, 2017. https://www.2ascribe.com/transcription-services/radiology-dictation-transcription.
97. Liu Y, Wang J. PACS and digital medicine: essential principles and modern practice. 1st ed. Boca Raton: CRC Press;2010.
98. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015; 372:793–795.
99. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014; 5:4006.
100. Hesketh RL, Zhu AX, Oklu R. Radiomics and circulating tumor cells: personalized care in hepatocellular carcinoma? Diagn Interv Radiol. 2015; 21:78–84.
101. Scikit-learn.org Web site. Scikit-learn algorithm cheat-sheet. Accessed April 1, 2017. http://scikit-learn.org/stable/tutorial/machine_learning_map/.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download