INTRODUCTION
Glioblastoma multiforme (GBM), the most frequent and malignant brain tumor in adults (
1), continues to show poor prognoses and low survival rates despite decades of multimodality treatment research. Along with the increasing availability of genomic data for the brain tumors, active studies are underway to enable genomic characterization and improved clinical outcome of GBM patients. Recent studies have revealed the genomic characteristics of GBM including the distinct patterns in gene expression profiles, underlying genomic abnormalities, and epigenetic modifications (
2). Improved understanding of the behavior of GBM at the molecular and genomic levels could lead to development of new drugs as well as patient-specific treatment regimens, thus facilitating precision medicine in the clinical field.
As knowledge of GBM increases from the genomic and clinical perspective, there is a growing need for reliable and efficient extraction of quantitative features from multimodality imaging data for associating imaging tumor phenotypes with genomic characteristics as well as clinical prognosis.
Radiomics, which refers to the high-throughput extraction of a large amount of quantitative features from radiologic images, has emerged as a significant research interest across a variety of specialties (
345).
Several studies have shown the positive potential of radiomic features for treatment monitoring and outcome prediction as well as associating imaging phenotypes with genomic profiles in various tumors (
3467). For example, Aerts et al. (
3) have shown that proper analysis of radiomic features could lead to identification of signature features, which was effective in decoding of tumor phenotypes and predictive of patient prognosis in lung and head-and-neck cancer.
Glioblastoma multiforme has frequently been the study subject of radiomic and radiogenomic research. Diehn et al. (
8) have identified a set of MR imaging features highly associated with gene expression patterns of several well-known gene programs and predictive of overall survival in GBM patients. Zinn et al. (
9) have used a semi-automated segmentation technique to derive a set of volumetric features from MR images, and were able to produce radiogenomic mapping of edema or cellular invasion phenotypes in GBM. The association of imaging phenotypes with clinical outcomes as well as molecular subtypes, and related biological pathways in GBM (
1011121314) has been studied using MRI features derived from visual grading method, manually-drawn region of interest (ROI), or various types of computer-assisted techniques.
These studies have stressed the positive potential of the radiomic approach; however, evaluating the reliability of radiomic features in GBM is also important. Previously, lung cancer-related radiomic studies have evaluated the reliability as an integral part of study (
34), whereas the reliability of features in GBM remains unclear.
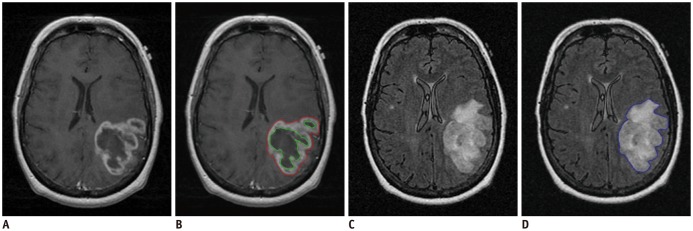
Tumor segmentation is regarded as the major source of variability in radiomics, since radiomic features are routinely derived from the segmented tumors using a computer algorithm (
13). In studying radiomics of GBM, tumor segmentation involves more complex tasks since differing tumor imaging phenotypes appear differently such as contrast enhancement, necrosis, and edema depending on MR sequences; in addition, image registration is required prior to tumor segmentation. Thus, tumor segmentation in GBM is prone to additional sources of variability and increases the uncertainty of feature reliability, which may lead to false positives if highly variable features were employed unknowingly. Therefore, evaluating the quality of radiomic features in GBM is an important and necessary step before translating into clinical application.
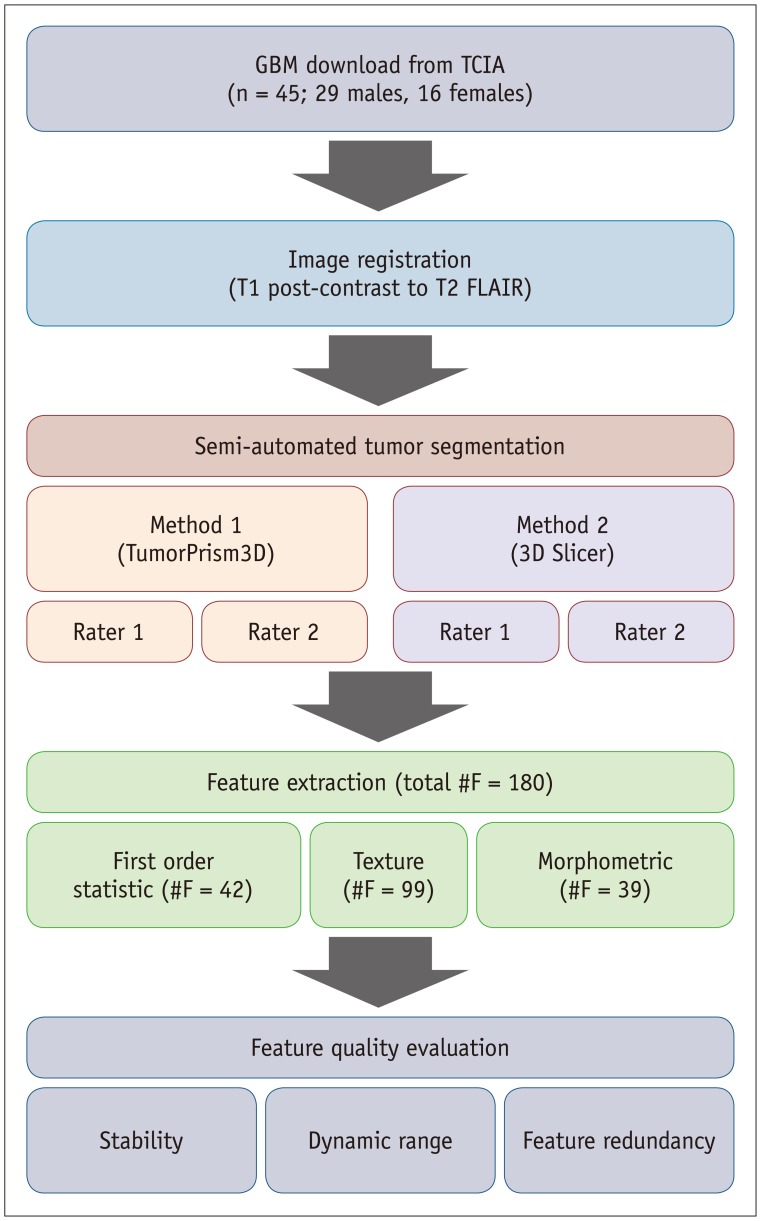
In this study, we evaluated the reliability of radiomic features in GBM derived via a computer-assisted tumor segmentation procedure. In particular, we assessed the feature stability against perturbations in tumor segmentation caused by varying raters and semi-automated segmentation techniques. In addition, we evaluated the normalized dynamic range (NDR) and redundancy of feature values thus qualifying the radiomic features in GBM in multiple aspects.
DISCUSSION
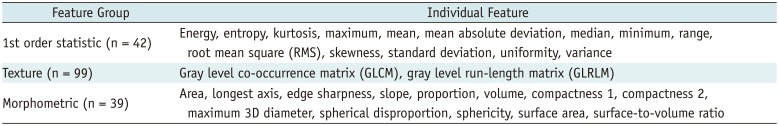
In this study, we investigated the quality of radiomic features in GBM. We categorized radiomic features into first order statistic, texture, and morphometric feature groups and assessed their quality in terms of stability, NDR, and redundancy.
Image segmentation is the first step in radiomic feature analysis, and thus can be considered as a major source of variability in radiomics. Use of a semi-automated software for tumor segmentation is a preferred option in radiomics studies since it offers markedly improved efficiency and may reduce inter-observer variability in tumor delineation (
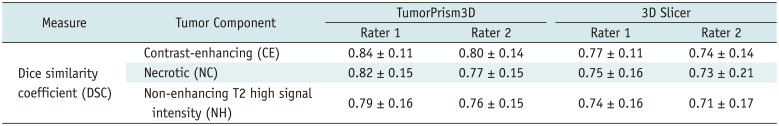
2324). The 3D Slicer has often been used in previous studies for segmenting 3D tumor volume due to its free availability in public domain. However, study settings may include different software tools employing differently evolved algorithms. Therefore, comparing the performance of different semi-automated software tools in terms of quality assessment of radiomic features could form the basis for study design and experimental tools.
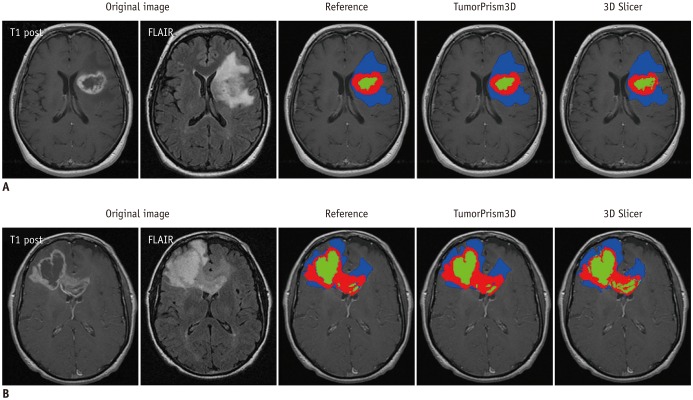
In our experiment, using two semi-automated software tools, the tumor segmentation accuracy as assessed with DSC ranged from 0.71 to 0.84 depending on differing raters and software tools. Compared to reported values of segmentation accuracy of brain tumors that ranged considerably (0.48 to 0.97) (
25), the two software tools employed in our study appear to provide consistently good accuracy in segmenting GBM tumors. Thus, we regarded the two software tools as adequate for use in the subsequent assessment of the quality of radiomic features in GBM.
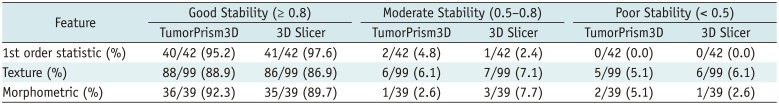
Stability has been often used for quality assurance and selection of robust features at the first step in radiomic feature analysis (
4). Our study results showed that most of the radiomic features in GBM were highly stable. Over 90% of 180 features showed good stability (ICC ≥ 0.8), whereas only 7 features had poor stability (ICC < 0.5) with both software tools. In general, first order statistic group showed relatively higher stability, followed by morphometric group and texture group, in order. These results agree with the data reported by Parmar et al. (
4). They examined the stability of radiomic features in CT lung cancer scans against three independent raters with the 3D Slicer as a semi-automated segmentation tool; the results indicated overall high ICC (0.85 ± 0.15) with 74% of 3D radiomic features showing good stability (ICC ≥ 0.8) and only 3 of 56 features showing poor stability (ICC < 0.5). This congruence suggests that these semi-automated software tools are sufficiently reliable in extracting radiomic features in different study settings, including CT for the diagnosis of lung cancer and MRI for the diagnosis of GBM.
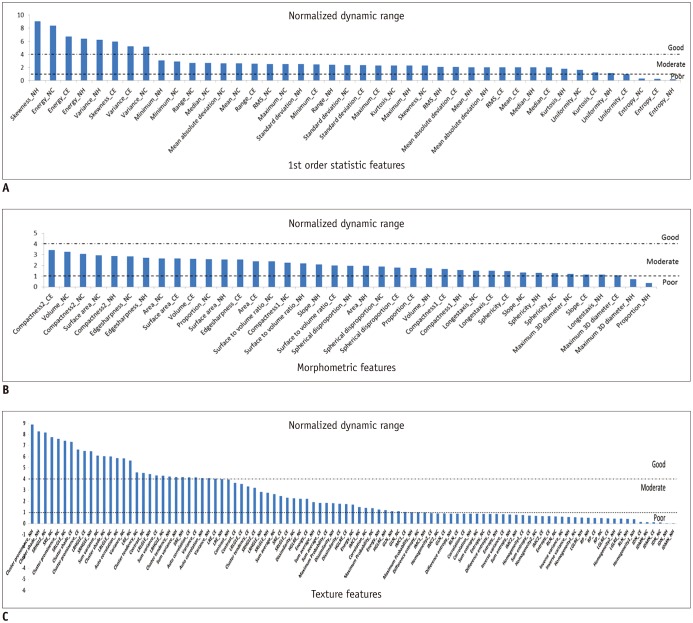
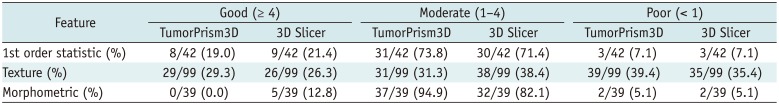
Dynamic range has often been used as a measure of informativeness of radiomic features. As a certain degree of perturbation is unavoidable in extracted radiomic features due to inherent variability from different sources, features with higher dynamic range are regarded as more robust to feature perturbation and thus regarded to possess relatively good information compared to those with narrow dynamic range.
In this study, we defined the NDR as the dynamic range of a feature over the study population divided by its mean; NDR was used for comparison of the relative dynamic range of radiomic features regardless of their feature value range. Our study results indicated differences in NDR among differing feature groups. Most first order statistics and morphometric features (93 and 95%, respectively) showed good or moderate NDR. In contrast, texture features showed relatively lower NDR, with > 35% of texture features of poor NDR. In a previous study on reproducibility of radiomic features in CT lung cancer, Balagurunathan et al. (
26) examined the dynamic range of different feature groups, and also found that significant differences existed in dynamic range of different features.
Majority of the morphometric and histogram features were among the higher rank of dynamic range; whereas, significant portion of texture features were among the lower rank. These findings suggest the need for an understanding of differences in feature dynamic range in determining truly reliable and informative features in radiomic feature analysis.
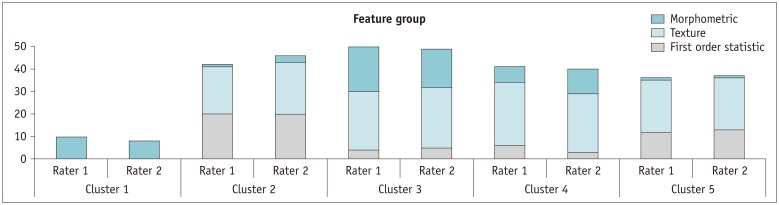
Typically, several features are extracted at an early stage in the radiomic feature-analysis process, which may reach up to several hundreds and exceed the number of samples. This can cause data overfitting and increase the risk of false positives leading to decreased reliability of the study results. Therefore, reducing the redundant features and choosing a small subset of representative signature features is an essential step in radiomics study. In this process, assessment of feature cluster property reflects the degree of similarity as well as distinctness among feature groups and thus facilitates the determination of the characteristics of a representative feature subset.
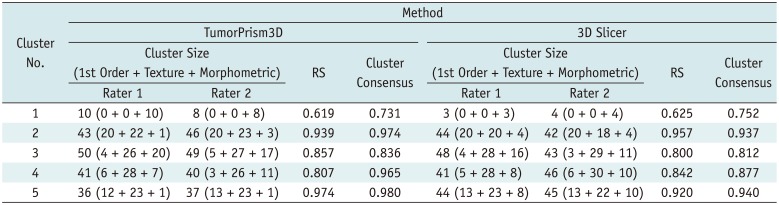
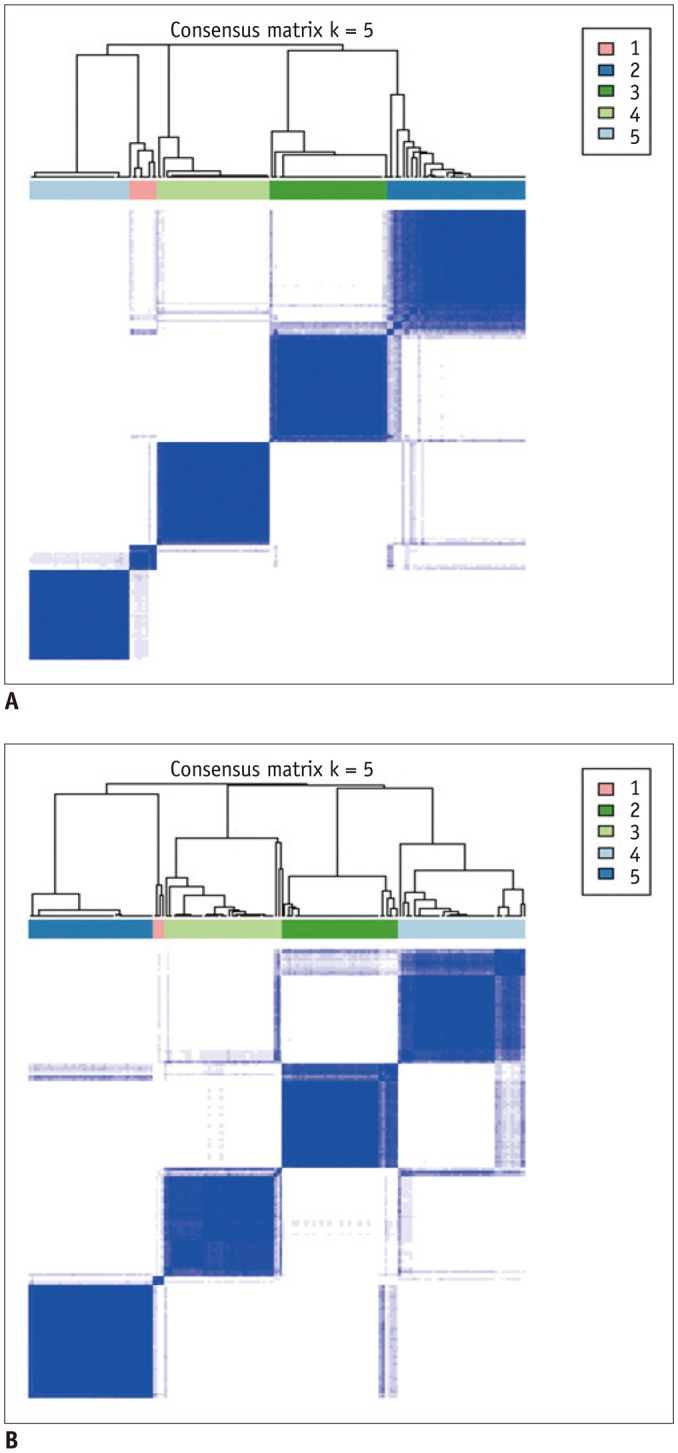
The CC map produced in our study revealed that the 180 features were highly redundant and could be compressed into 5 distinct clusters. In addition, both CC and RS derived using two different segmentation software tools showed very similar trends across the five clusters. These findings suggest that those feature cluster properties shown in our study were of fundamental nature in radiomic features of GBM regardless of segmentation software tools and rater's experience.
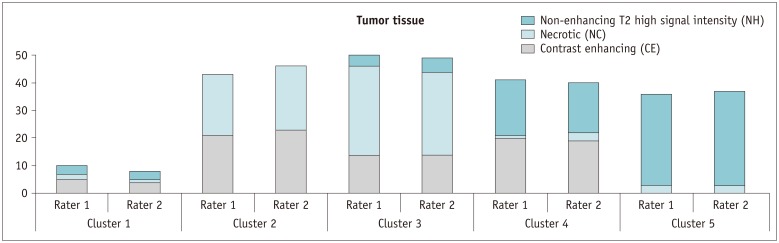
We expected that diverse delineation pattern of CE, NC, and NH tumor tissues appearing on multi-parametric MR images would form more complex cluster pattern. However, we identified 5 clusters, which was less than the previously reported 11 and 13 clusters found from 440 CT radiomic features of lung cancer and head and neck cancer, respectively (
27).
Significant differences existed in the proportion of features based on the concurrent tumor components. A substantial proportion of features (in clusters 2 and 3) related to CE and NC tissues, which might indicate a strong interaction between the CE and NC components. In contrast, less proportion of features were of CE and NH tissues (in cluster 4), which might indicate a weaker interaction between the CE and NH components. Further study is required for a pathophysiological interpretation of this finding.
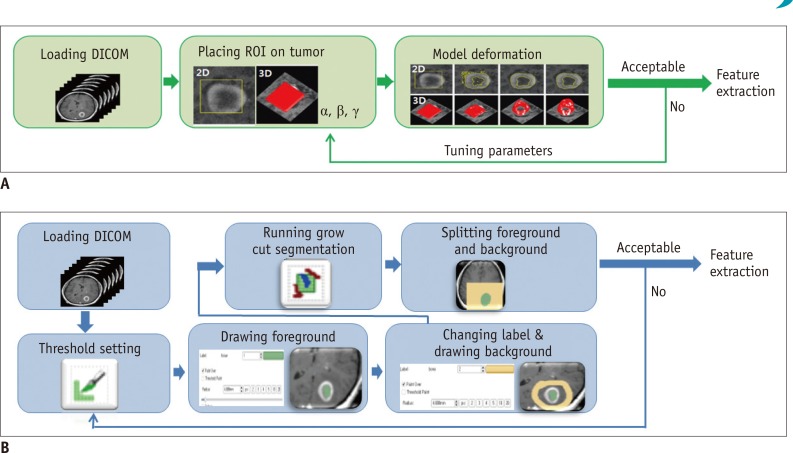
Our study has several limitations. First, only two types of software tools were used for evaluation. Semi-automated segmentation tools employ sophisticated algorithms to reduce user's manual intervention as much as possible and provide reliable segmentation results at the same time. Computer vision community has developed different kinds of segmentation algorithms specialized for use in medical imagery. To our best knowledge, grow-cut algorithm and deformable model-based algorithm were two representative semi-automated algorithms for segmenting tumors on medical images, and were implemented in the 3D Slicer and the TumorPrism3D, respectively. As image segmentation algorithms continue to evolve requiring less intervention from users and making use of more learned knowledge from a large image database, radiomics applications of additional software tools with novel algorithms should be investigated in future.
Second, stability of radiomic features was evaluated with a single scan dataset. Though variation of segmented tumor volumes due to inconsistent user intervention is regarded a major source of instability of radiomic features, varying physio-physical state of patient and scanner might cause additional perturbations to radiomic features. Accordingly, it would be desirable to use a same-day repeat scan data set to evaluate the stability of features against overall sources of variability. However, such a data set was not available to our study. Therefore, our stability data should be interpreted with caution in that they are applicable only to limited sources of variability.
In addition, our study used an MRI data set acquired with a relatively simple protocol. As a variety of pulse sequences are used in MR imaging, it is obvious that images acquired with different pulse sequence would bring about different feature quality in GBM study. For example, diffusion imaging is increasingly used in GBM, which produces much noisier images and accordingly would give rise to radiomic features of much different quality. Thus, our study results cannot be generalized to all MR GBM studies.
In conclusion, the use of two different semi-automated software tools by different raters showed similar high stability in radiomic features in GBM regardless of difference in raters' experience, indicating that semi-automated software tools provide sufficiently reliable segmentation output and help overcome the inherent inter-and intra-rater variability from user intervention. However, significant differences existed in NDR among features, which suggests that features convey information with differing strength. Among the feature groups, texture features showed the weakest NDR. A total of 180 radiomic features in our study were highly redundant, and compressible to 5 distinct clusters. However, significant differences existed in the proportion of features according to the tumor tissue components appearing together.
A well-established quality assurance procedure has an important role in the future advancement of radiomics and translation into patient care. The findings in our study may be useful in guiding the development of quality assurance procedure of the radiomics pipeline, particularly for GBM.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


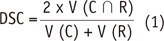
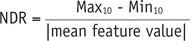
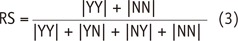
 XML Download
XML Download