Abstract
Within six months of the discovery of X-ray in 1895, the technology was used to scan the interior of the human body, paving the way for many innovations in the field of medicine, including an ultrasound device in 1950, a CT scanner in 1972, and MRI in 1980. More recent decades have witnessed developments such as digital imaging using a picture archiving and communication system, computer-aided detection/diagnosis, organ-specific workstations, and molecular, functional, and quantitative imaging. One of the latest technical breakthrough in the field of radiology has been imaging genomics and robotic interventions for biopsy and theragnosis. This review provides an engineering perspective on these developments and several other megatrends in radiology.
Go to : 
Within 6 months of Röntgen's discovery of X-ray in 1895, physicians were already utilizing the resulting technology to diagnose and treat diseases (1). Nonetheless, there were still many barriers to achieving its full use. In October, 1920, 13 technicians established the American Association of Radiological Technicians (AART), the first national society of its kind, to overcome these obstacles by sharing ideas and skills on radiologic techniques (1). Since the founding of the AART, radiology has developed rapidly and plays an increasingly important role in the diagnosis of diseases in various clinical settings.
In the recent decades, there have been many new radiologic imaging modalities, including ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI), with numerous applications to clinical practice. The first CT device was invented in 1972 by British engineer, Sir Godfrey Hounsfield, who worked at Electronic & Music Industries Ltd., and South African physicist, Allan Cormack, of Tufts University. The first clinical CT scanners were installed in Atkinson Morley Hospital in Wimbledon, England, and the first patient brain scan was performed on October 1, 1971 (2). In 1979, 7 years after the announcement of their invention, Hounsfield and Cormack were awarded the Nobel Prize for their contributions to medicine and science.
Beyond developments in the types of imaging modalities, there have been additional megatrends in radiology that are of equal importance, including 'being digital,' new display and input devices for picture archiving and communication system (PACS), 3D intelligent and organ-specific workstations, functional and quantitative imaging for imaging biomarkers, imaging genomics, bionics, and robotic interventions for interventional radiology. The digitization of medical imaging allows unlimited data storage and sharing without substantial effort. Spiral CT and volumetric MRI enable 3D visualization and quantitative analysis at the diagnostic workstation, which has become organ-specific in order to more effectively respond to the clinical need at hand. At the present time, the digital state demands that decision support systems based on clinical knowledge modeling, such as computer-aided detection/diagnosis (CAD), are seamlessly integrated into PACS. In addition, the introduction of new display and input devices, including stereovision, holograms, 3D mouse, and stereo cameras, are expanding the ability of the radiologist, who can now efficiently deal with very large amounts of radiologic information.
Molecular, functional, and quantitative imaging is an increasingly important capability in the field of radiology. Oncologic imaging is the clearest example in which functional and quantitative image metrics such as tumor diameter, tumor volume, standard uptake value, and permeability must be determined to assess treatment response. These functional and quantitative data, accessed by the interpreting radiologist in many cases using automated software tools, yield more definitive and ultimately more accurate diagnostic conclusions. From an engineering point of view, the final objective is to establish processes and profiles of the following, which, are accepted by the imaging community, the clinical trial industry, and regulatory agencies: imaging biomarkers as proof of biology, indicators of alterations in the underlying pathophysiology, and surrogate endpoints for changes in the health status of patients.
Imaging genomics is very likely to become another megatrend in radiology. The recent completion of human genome sequencing promises to provide unprecedented opportunities to explore the genetic basis of individual differences, which specifically requires radiologic imaging devices. These may offer a complementary strategy in the examination of genotype-phenotype relationships. Deciphering the complex nature of these relationships will require sophisticated computational methodologies.
The megatrend toward minimally invasive image-guided interventions has also created new challenges in the introduction of robotic technology. For example, the use of master-slave robot control technology and accurate pre-planning support may reduce radiation exposure to operators and patients. In addition, micro-imaging devices and various sensors will improve the safety of image guided interventions.
In this review, we introduce many of these promising megatrends in radiology, i.e. those with the potential to change the future of radiology, and provide insight into their applications from an engineering perspective.
Go to : 
Unlimited data storage and sharing capabilities, which have rapidly become routine, reflect the digitization of medical imaging (Fig. 1). Consequently, PACS solutions have become an important component of radiology departments, with respect to the infrastructure responsible for medical image management according to the well-known standards elaborated in Digital Imaging and Communications in Medicine (DICOM) (3), Integrating Healthcare Enterprise (4), and Health Level Seven International (5). Since most imaging modalities transmit their images through these standard routes, all such images can be stored in a single PACS solution and then retrieved by physicians using their own desktop imaging software. Although PACS solutions are a vital and well-established commodity in diagnostic radiology, they are still expected to provide an innovation platform in radiology (6). Thus, regardless of the many functions already integrated within state of the art PACS solutions, upgrades by the major vendors continue to enhance productivity relying on medical images based on the following megatrends:
Firstly, PACS is now expanding the territory of its applications beyond the radiology department. Many clinical departments, such as cardiology, dentistry, and radiation therapy, are incorporating medical imaging modalities into their workflow (7). Recently, the DICOM standards have been extended to incorporate medical specialties, such as radiotherapy, cardiology, pathology, and ophthalmology, such that the images can be viewed with respect to their specialty-specific information (8, 9). This has necessitated a wide range of modifications in the pre-existing radiology PACS, since each specialty has its own specific workflow, in addition to the fact that the primary nature of the various imaging modalities differs. So far, most large hospitals have implemented separate and independent PACS solutions for each department, although this is an unnecessarily complex and inefficient way to share images. Instead, it would be preferable for one given PACS solution to manage all patient-relevant images in a unified manner, with server-side components and storage commonly shared by all departments, and optimized access for client viewers according to the requirements of each clinical department.
Secondly, PACS viewing is adopting a growing number of image processing algorithms and 3D/4D visualization technologies to support the intuitive and quantitative analyses of clinicians. Nowadays, most PACS are equipped with the advanced visualization and image processing algorithms that, until recently, were available for high-end workstations only. For example, doctors can use PACS with CAD to detect or diagnose suspicious regions more easily and productively (10). Such functions must be available within the PACS viewer itself, as separate workstations are unable to significantly enhance daily routine workflow, due to the computational complexity, network bandwidth limitation, and the lack of hardware capabilities at the client workstations. To overcome these obstacles, server-side computing technologies, such as the thin-client approach, have been used intensively for PACS implementation (11). Transmission of the images to the central PACS solution allows its computing machines to be configured, such that computer-intensive image processing is performed automatically. Doctors can then view the pre-processed results, including those related to segmentation, registration, quantification, or CAD. In addition, thin-client technology will enable doctors to make use of the full range of complex image processing and visualization techniques anywhere and at anytime. These technologies will be the key foundation in a mobile environment, in which the development of devices such as smartphones and tablets, including Google Android™ devices, Apple iPhone™, and iPad™, are already recognized as one of the most significant developments in the IT world.
Go to : 
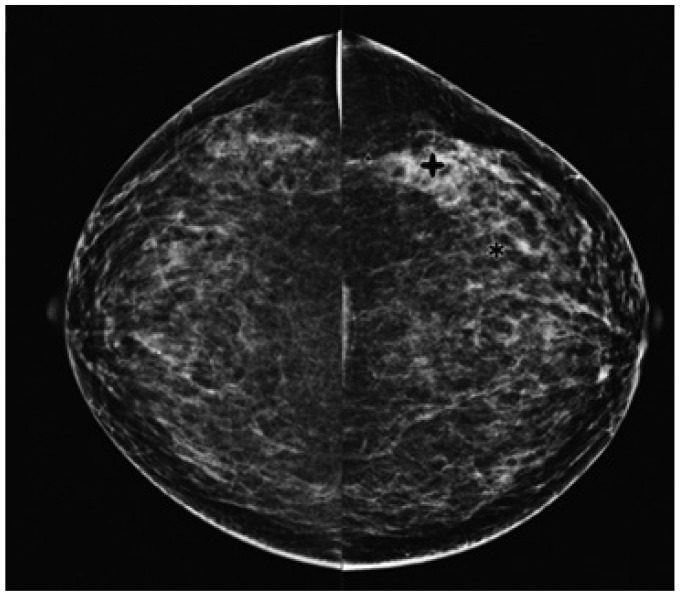
Clinical intelligence and knowledge must be integrated into PACS solutions to support daily radiology tasks. Although attempts at computerizing the analysis of medical images (12, 13) were previously made in the 1960s, serious and systematic investigations into the possibility of CAD began in the 1980s, paralleling a fundamental change in the concept of computer output utilization, from computer-automated to computer-aided diagnosis (14-17). While those efforts were initially hindered by the difficulty of digitalizing medical images (14), CAD has since become a major focus of research, and developments in this field have been incorporated into the routine diagnostic radiology approach to the detection of breast cancer on mammograms (Fig. 2) (18-24). The motivation and philosophy guiding the early development of CAD was the application of intelligence and knowledge to PACS, based on medical images being digital. With CAD, radiologists are able to use computer decision supports as a second opinion in reaching a final diagnosis, which means that computer performance compliments decision making by physicians. In fact, in addition to the aforementioned application in breast cancer detection (18-23), CAD has the potential to improve overall performance in the detection of lung nodules (25-29) and vertebral fractures. The latter can be reliably detected by CAD on lateral chest radiographs (30), thus improving the early diagnosis of osteoporosis (31). In MR angiography, a CAD system has been developed to detect intracranial aneurysms (32, 33). Interval changes on successive bone scan images, such as those obtained from patients with suspected bone metastases, can also be detected using a CAD system, based on the subtraction images (34).
Many different types of CAD systems have been recently implemented as part of PACS solutions (10, 35-38). In chest radiographs and CT scans, the package for chest CAD might include the automated detection of lung nodules, interstitial opacities, cardiomegaly, vertebral fractures, interval changes, etc., as well as the automatic differentiation of benign and malignant nodules, and the differential diagnosis of interstitial lung diseases. This seamless integration of CAD and PACS significantly increases reader sensitivity and reduces the average image reading time, thus vastly improving the efficiency of daily clinical practice (35). In the near future, it should be possible to use PACS to search for and retrieve lesion images that are similar in appearance to previously queried images, based on a set of primitive image characteristics which are not related to any specific diagnostic method able to visually characterize the image, by utilizing reliable and practical methods developed for quantifying the similarity of a pair of images for further visual comparison by radiologists (39-42). Similar content-based image search engines will no doubt be of interest to all medical specialties that make use of medical imaging or digital biomedical signals, drawing support from the information available in medical archives (40).
Go to : 
Beyond the simple display of medical images, radiologic diagnosis demands more specialized image processing, based on targeted organ types, organ-specific diagnosis, or organ-specific surgery procedures, in order to increase the accuracy and the efficiency of the workflow. Therefore, not only major modality vendors (Siemens Healthcare, Erlangen, Germany, GE Healthcare, Milwaukee, WI, USA, Toshiba Medical Systems, Tokyo, Japan, and Philips, Best, The Netherlands) (Fig. 3), but also 3D medical image visualization and processing software companies (e.g., TeraRecon, MeVis, Aze, and INRIA) have been expanding their 3D visualization platforms to include organ-specific workstations. In particular, Aze has developed organ-specific workstations with more than 50 organ-specific modules, while software companies such as EDDA Technology and Diagnostics focus on specific organs, such as liver and lung, respectively. The organ-specific capabilities of these workstations are detailed as follows.
Cardiology workstations differ from general radiology workstations. In the latter, the workflow for viewing and reporting study cases is primarily driven by an imaging request, whereas the cardiology workstation must access all cardiology imaging data for each patient, which may be stored across a number of different storage systems. Thus, data recorded during a cardiac catheterization procedure would also be available for viewing on a cardiology workstation.
Brain-imaging workstations require advanced visualization techniques to allow the interpretation of images from the various MRI imaging (anatomic, perfusion, diffusion, functional, etc.). Some commercial stand-alone workstations, e.g., BrainVoyager and BrainMagix, are specifically devoted to brain imaging. In addition to commercial workstations, a number of academic open source programs have been developed and distributed for various purposes such as brain segmentation, functional imaging analysis, and diffusion tensor imaging (DTI). Well-known systems include Harvard University's FreeSurfer, for the analysis of the cortical thickness of the brain (43, 44); the University of Birmingham's Brain Imaging Lab, for general neuroimaging analysis (45); and the University of Pennsylvania's DTI-TK, for atlas construction (46).
Mammography also requires a workstation with a specialized functionality, because the ability to detect fine details in mammography images is vital to their effective use in diagnosis. Therefore, image analysis software must be capable of resolving micro-calcifications, the appearance of which can be a clinical indicator of pre-cancerous changes in the breast. Currently, there are several commercial systems that offer the required functions, such as IDI Mammography Workflow Solutions (GE Healthcare, Milwaukes, WI, USA), IDS7/MX (Sectra: Linköping, Sweden), and the BX Mammography Workstation (Neusoft Neusoft, Shenyang, China).
Finally, lung workstations also require special processing algorithms, such as lobar segmentation, airway segmentation, airway tree labeling and wall measurement, parenchymal density analysis, and emphysema analysis. The pulmonary workstation developed by VIDA Diagnostics has been widely adopted. The GE Advantage Workstation™ has advanced lung analysis modules, while the TeraRecon Aquarius workstation includes lung nodule tracking and analysis functions. In addition, some academic labs in South Korea and Netherlands have developed in-house software for lung specific workstations (47-55).
Go to : 
Despite the many advantages offered by these technologies, the differential diagnosis together with the standard qualitative reporting procedure is very likely to remain the heart and soul of radiology. However, radiology is gradually embracing functional and quantitative metrics, which provide vital information in an increasing number of radiology settings.
In general, functional imaging means functional magnetic resonance imaging (fMRI) to detect blood-oxygen-level-dependent contrast material, as an indicator of brain neuronal activity. Following the groundbreaking discovery of fMRI by Ogawa et al. (56), it was applied in a large number of studies aimed at discovering the 'secrets' of the brain. Thus far, fMRI has been used to study not only neuroscience with respect to memory, cognition, brain-robot interfaces, etc., but also to obtain evidence of the diagnosis-specific patterns of brain activation in various neurologic diseases (57-61).
The management of oncology patients increasingly depends on medical imaging to reach a diagnosis, as well as to monitor treatment response and follow-up. The clinical applicability of combined functional and anatomical imaging modalities, which integrate the benefits of visualizing tumor biology with high-resolution anatomical imaging, has revolutionized the clinical management of cancer patients (62). High-resolution anatomical imaging modalities such as CT and MRI provide detailed structural information regarding lesion location, size, morphology, and morphological changes, but they do little to further the understanding of tumor physiology. With the increasing focus on molecularly targeted therapies, imaging radio-labeled compounds with positron emission tomography (PET) and single-photon emission tomography (SPECT) are often carried out to gain insights into the biology and surrounding environment of the tumor (63, 64). The availability of multimodality imaging with PET/CT, SPECT/CT, and PET/MRI has the potential to improve lesion characterization, treatment decision-making, and patient management, to name just a few of its potential advantages. In addition, continual developments in instrumentation and imaging agents will improve the ability to non-invasively evaluate disease processes (64).
Based on these developments, the Radiological Society of North America (RSNA), has committed to help transform patient care by making radiology a more quantitative science (65-68). Quantitative imaging is the extraction of quantifiable information from medical images to define the normal condition or to measure the severity, degree of change, or status of a disease, injury, or chronic condition relative to normal. This requires the development, standardization, and optimization of anatomical, functional, and molecular imaging acquisition protocols, data analyses, display techniques, and reporting systems. This information will allow the validation of accurately and precisely obtained image-derived numerical metrics with anatomically and physiologically relevant parameters, including treatment response and outcome, and the use of such metrics in research and patient care.
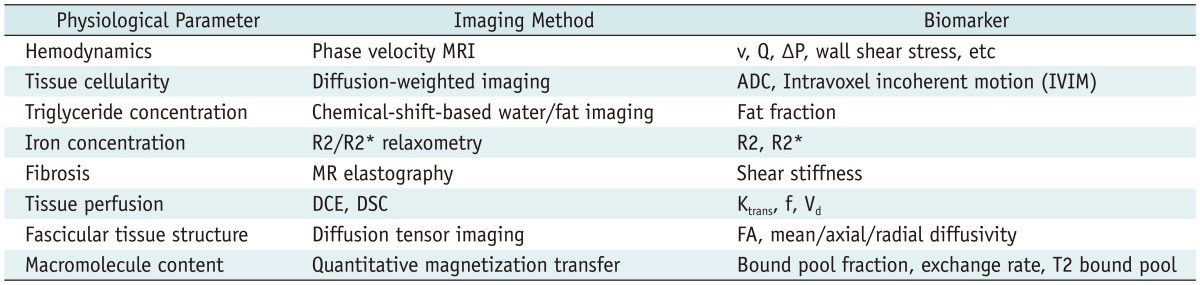
Efforts to accumulate public resources and open source tools in order to qualify longitudinal volumetric CT imaging for use with imaging biomarkers (Table 1), i.e., reproducible biologic features detectable by imaging modalities (69), were re-invigorated in 2005 by an informal alliance between the US Food and Drug Administration (FDA), the National Cancer Institute, the National Institute of Standards and Technology (NIST), and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) (70-74). The preliminary initiative of this inter-federal agency led to the organization of a public workshop, which was held at the NIST headquarters in 2006 and modeled on the Integrating Healthcare Enterprise. The Scientific Advisory Board of the RSNA met in 2006 and consequently established a Quantitative Imaging Biomarker Alliance (QIBA), with the aim of advancing quantitative imaging and the use of imaging biomarkers in clinical trials and clinical practice by engaging researchers, healthcare professionals, and industry members (67). RSNA was awarded a two year, $2.4 million contract from the NIBIB to support QIBA. This contract encouraged a coordinated effort to establish an infrastructure for collecting and analyzing imaging biomarker data. The purposes of QIBA are two-fold: collaborating to identify needs, barriers, and solutions to develop and test consistent, reliable, valid, and achievable quantitative imaging results across imaging platforms, clinical sites, and time; accelerating the development and adoption of hardware and software standards needed to achieve accurate and reproducible quantitative results from imaging methods (75). In terms of engineering, QIBA could resolve major problems, including differences between vendors or between different machine versions of the same vendor.
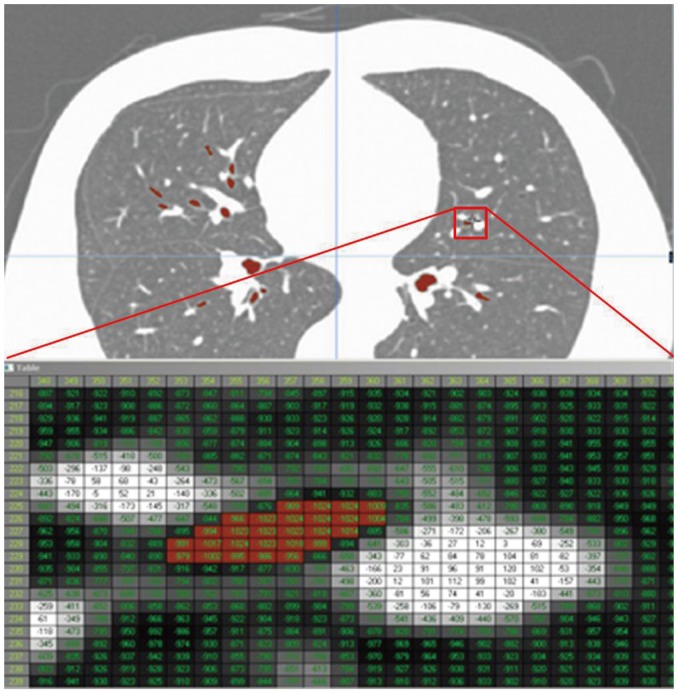
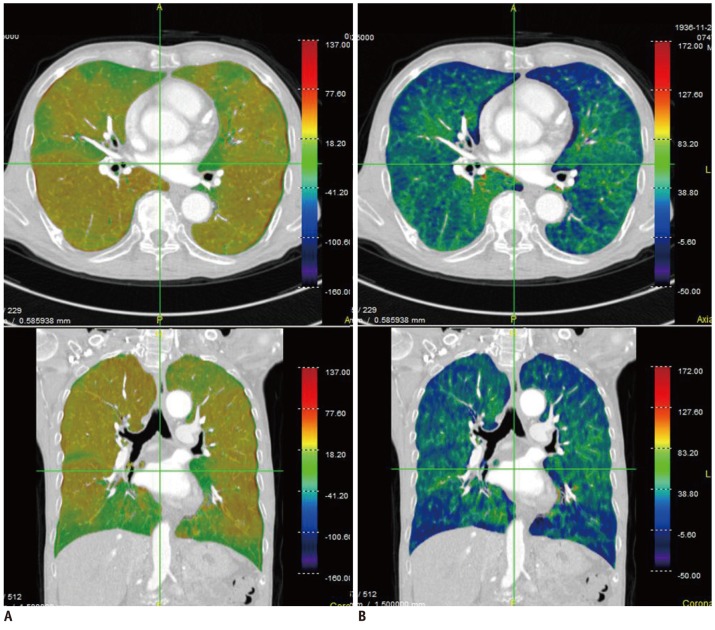
Extensive research has also been carried out in the field of pulmonary functional and quantitative imaging. In the former, xenon ventilation (76, 77) and iodine perfusion (47) using dual-energy CT have been evaluated (Fig. 4). In quantitative imaging, lung segmentation, airway measurement, textural analysis of the parenchyma, automatic quantification, follow-up analysis, and clinical applications were examined (47-53).
Go to : 
With the goal of diagnosing and evaluating patients non-invasively, medical visualization is a highly intuitive approach, and its applications are being continually extended. Effective visualization of the disease region to determine its position is likely to go beyond the realm of diagnosis to include minimally- or non-invasive examination and treatment. The emerging technologies to display the products of visualization and the input devices for seamless interaction with users are introduced in this section.
One of the trends of display devices is to show more realistic depictions by making use of 3D depths, such as stereovision (78). When combined with a 3D diagnostic workstation, surgical planning software, simulation, and a robot-master console, there are benefits in terms of usability, as relative depths are displayed among the complex 3D anatomical shapes, the different organs, and the end effectors.
In general, while most stereovision systems require special glasses, 3D virtual scenes can be generated without them if the user's typical viewing distance and viewing direction are within the specified range, such as is demonstrated by the small mobile devices (79, 80). Another option is holographic technology, which generates 3D objects rather than a virtual model. Pennsylvania based Ark Media shows a cylindrical holographic example of the lungs (81) that can be viewed 360° (82).
While realistic virtual reality or holography has many technical limitations, augmented reality offers greater opportunities and feasibility. In the operating room, separate 2D monitors are generally used to show pre-operative and intra-operative medical images, with the positions of current surgical devices obtained via surgical navigations. However, viewing distracts visual attention from the surgical field to the display monitors, thus disturbing the surgeon's concentration and requiring a substantial amount of imagination and attention in the interpretation of the displayed images. Accurately and effectively displaying the image directly on the patient's body would obviate the need for a mapping process between the monitor and the patient. These technologies could be useful to the interventional radiologist in determining entry points and the direction of needle insertion.
In this context, Kutter et al. (83) developed a real-time volume-rendering system that is set up over the patient's body and makes use of the head mount display devices shown, while Werkgartner et al. (84) explored a 3D model overlay on a 2D image plane. Although their clinical applicability awaits further testing, these novel systems may provide an alternative method, allowing intuitive and seamless access to displays in the operating room.
Another trend in display devices is portability. High-resolution portable devices and thin-client software systems via networks allow diagnoses to be made on demand. The limited size of the mobile display devices can be solved by small pico-projectors such as the one shown in Figure 5.
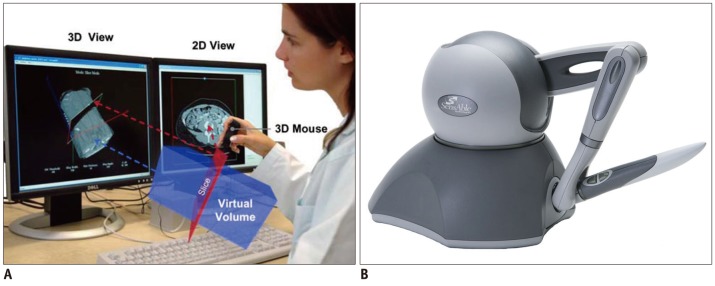
As the popularity of 3D display devices continues to grow, the requirements for new, compatible input devices will also grow. Figure 6 shows 3D input devices allowing the control of medical images and a haptic device. A 3D mouse is an input device with more than 3 degrees of freedom (DOF) about a rigid-body transformation. Typically, there are 6 DOF: 3 related to translation, and a maximum of 3 related to rotation. The 3D mouse shown in Figure 7A is used to interact with the medical image browser (85). Sometimes haptic devices that provide force feedback, including 6 DOF positioning and remote robotic control, are used for special purposes.
In reality, as opposed to virtual space, stereo acquisition systems are generally used to obtain depth information. Similar to human vision, the depth information can be calculated using the difference perceived by binocular vision. Figure 7B shows a low-priced general-purpose stereo camera, Kinect™, developed by Microsoft Corp. Since its software development toolkit includes a human motion analysis function, a visual sensor for the detection of human gestures has attracted attention. However, the current version is plagued by the problems of latency and relatively large spatial errors when used for images. For fast and accurate 3D tracking, special markers are often adopted, as shown in Figure 7A. These have been widely employed in medical applications as a navigation system.
Go to : 
Imaging genomics is defined as an association analysis between genes and a physiological response of the brain during specific information processing, which is captured in images (86). It is a field of study that integrates molecular genetics, represented by the Human Genome Project, with neuroscience, based on recent progress in neuroimaging modalities, since approximately 70% of all genes are expressed in the brain (86). Closely related to bioinformatics, imaging genomics has advanced rapidly because of the increase in scanning data obtained from individuals using structural and functional MRI, and the emergence of voxel-wise methods that search every location in the brain for statistical analysis (87, 88).
Many genes expressed in the brain are important in disease, including behavioral disorders, not only with respect to understanding disease mechanism but also in the diagnosis of patients with suspected pathologies, as well as in the identification of individuals at risk and in the development of new treatments. Even if genes are not solely responsible for disease, twin studies have shown 40-70% heritability in cognition, personality, and other human behavioral patterns (86). In addition, genes are known to be the only consistent risk factor for psychiatric disease. The 3D neuro-anatomical patterns of gene effects can be visually assessed using DTI.
Magnetic resonance imaging-based methods including DTI and fMRI are regarded as the most fruitful modalities in imaging genomics. In addition, there may be related applications that benefit from nuclear medicine modalities, i.e., PET and SPECT. While magnetoencephalography and electroencephalography are also of interest in this setting, this paper will focus on the various MRI sequences.
To apply neuroimaging to a specific behavioral study, Hariri and Weinberger (86) suggested three general procedures: selection of candidate genes, control for non-genetic factors, and task selection. The first step involves the selection of the appropriate candidate genes, that is, those related to the behavior of interest. Depending on the gene, functional polymorphisms may be well known, circumscribed, or unknown. While candidate genes with identified single nucleotide polymorphisms are tractable in imaging genomics, studies based on genes which have poorly or not at all understood functional effects and their variations should be carefully designed. Second, non-genetic factors must be controlled for, as the effects of a single gene on the brain may be limited. Thus age, gender, or environmental factors may prevent the effects of a single gene from being detected unless a carefully matched control is included across genotype groups. Lastly, as the effects of single genes are relatively small, and potential gene effects are the detection targets of information processing, it is recommended that imaging tasks that are known to be effective for information processing are selected in order to maximize the sensitivity of those tasks.
For example, apolipoprotein E4 (APOE4) is one of the most studied polymorphisms in neuroimaging. The various APOE4 alleles are known to increase the risk for developing late-onset Alzheimer's disease (before the age of 75 years) by three-fold. A decrease in the volumes of grey and white matter in the brains of the elderly is also related to APOE4 levels (89-91). Children carrying this gene later show cortical thinning and trajectory changes; in addition, resting-state activity in the young brains of these gene carriers is modified (92, 93). Besides genes related to Alzheimer's disease and schizophrenia, mutations of the Parkin gene (PARK2) and the PINK1 gene (PARK6) are known to cause Parkinson's disease. Van Nuenen performed a neuroimaging study for individuals with PINK1 (94). In tumor evaluation, the gene expression patterns for a mouse tumor model have been attempted using MRI (95).
Imaging genomics is a promising field of study that has many advantages over earlier approaches to genetic research, especially in light of the steady advances in medical imaging technologies. Imaging genomics may be better than epidemiological studies, as the former can demonstrate the roles of genes in psychotic disorders, including schizophrenia, and requires fewer subjects with high behavioral accuracies (96).
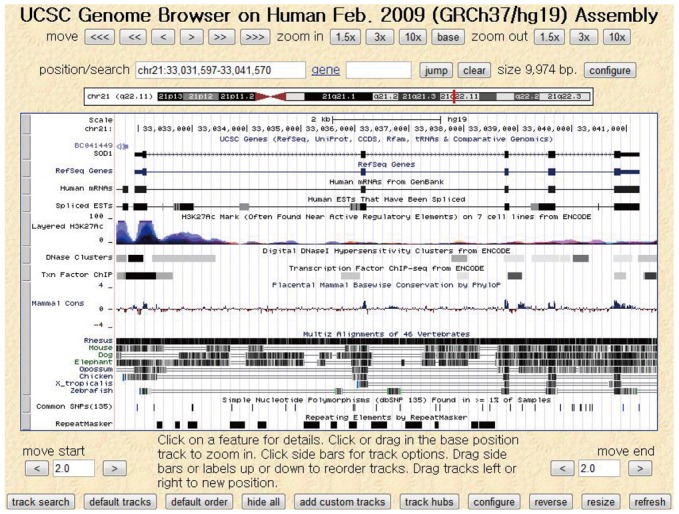
Active studies on imaging genomics began with the completion of the Human Genome Project in 2003 and are still ongoing (87). Many academic institutes have provided software, databases, and websites to facilitate gene studies. For instance, the University of California, Santa Cruz hosts an online genome browser service (Fig. 8), called the UCSC Genome Browser, in which users can access a database of genome sequences and download related documentation (97). Another example is the Galaxy platform, supported by multiple organizations including Pennsylvania State University and Emory University, which offers a scientific workflow, data integration, and analysis services with a graphical interface (98). In addition, international conferences such as the Imaging Genetics Conference at the University of California, Irvine continue to promote research on genes and medical imaging technologies. Imaging genomics has recently expanded from the brain to include the imaging of various organ-specific diseases, such as chronic obstructive pulmonary disease (99), cardiovascular disease (100), and cancer (101).
Go to : 
The word 'robot' was originally introduced in the play "Rossum's Universal Robot," written by the Czech dramatist Karel Capek in 1921, in which the meaning of robot was a laborer or servant (102, 103). It was in 1958 that robots were adopted in the real world for manufacturing automation, by the General Motors Company. Sometime later, robots began to be used in various fields.
The first medical robot was ROBODOC, released in 1992 by Integrated Surgical Systems; it was an automatic-drilling robot that inserted orthopedic implants in artificial-joint replacement surgery (104). However, as a fully automatic robot, obtaining FDA approval was a prolonged process which was not accomplished until 2008, at which point the company had partnered with a Korean company, Curexo Technology Corporation.
For laparoscopic surgery, human-controlled, master-slave robots were developed. In the 1980s, Scott Fisher carried out research in this field at the National Aeronautics and Space Administration (103). In the 1990s, Phil Green devised a tele-operated surgical system at Stanford Research Institute (SRI). Based on these developments, the US Department Of Defense invested tremendous amounts of money into the research and development of surgical robots (105, 106). Frederick Moll also acquired the technologies of a tele-operated surgical robot from SRI, and in 1995 he established Intuitive Surgical, which released the first version of the da Vinci surgical robot system (107, 108).
In this current technology, the surgeon's hand motions are used to manipulate robot arms and surgical instruments. Similar strategy has been applied to image-guided surgery, in which pre-operative or intra-operative CT/MR images are the basis for robot manipulation. With the development of the surgical robot, robotic intervention has been introduced, with remote manipulation systems for needle and catheter. Needle manipulation robots have a multi-joint actuation mechanism for instrument motion, which is controlled either by a computer that automatically analyses real-time images of the intervention or by a remotely located human operator with a master control device. This interventional real-time image can be obtained by ultrasound, X-ray fluoroscope, MRI etc. The motion of the master control device, guided by its human operator, is conveyed to the actual procedure slave robot. While needle manipulation robots have been intensively tested, they have not yet reached full-fledged commercialization, which means that more studies and innovative developments are required to prove their clinical efficacy. On the other hand, systems for catheter manipulation have been actively commercialized, and are drawing increasing interest in assisting catheter treatment. Meaningful success has been demonstrated both clinically and industrially.
Needle manipulation robots have a wide spectrum of clinical applications. As a general overview, the notable trends in their adoption can be summarized as follows. 1) Robots to assist needle biopsy under CT or X-ray fluoroscopy have recently re-gained research interest, possibly in accordance with the increasing interest in technologies for minimally-invasive treatments. 2) Numerous studies involving MR-compatible robotic biopsy are underway, in response to clinical demand for improvements in the diagnosis of prostate cancer. 3) One of the novel innovations in needle manipulation robots is steerable needle implementation.
In a few recent studies, active steering of the needle tip to achieve a curved insertion, in order to avoid vessels or dangerous regions, has been investigated. Nuebach and Shoham (109) reported a robotic system for flexible needle steering inside soft tissues, with real-time ultrasound imaging. An inverse kinematics algorithm based on a virtual spring model was applied to evaluate the needle base manipulations required for the tip to follow a predetermined curved trajectory. A closed-loop experiment with updated tissue stiffness parameters demonstrated a needle tip tracking error of less than 1 mm (109).
Recently, novel technologies have been explored by several research groups. Ganji et al. investigated the implementation of electromagnetic tracking of the spatial position of the catheter tip. The electromagnetic signal was used to achieve a kind of automatic control. The system automatically guides the motion of the catheter along a path specified by the operator (110). Patel et al. developed an actively controllable catheter, embedding a shape memory alloy. Through image processing and a visual-servo technique, the motion of the catheter is automatically controlled along a path determined by the visual signal (111). Plicchi et al. reported clear benefits for tele-control systems compared to conventional manual approaches, in that the operation time required for maneuvering the catheter is considerably reduced (112, 113).
Go to : 
Since the discovery of X-ray in 1895, radiology has opened the gate to new technical developments in the field of medicine, with a broad range of clinical applications. In this review, written from an engineering perspective, we have pointed out some of the megatrends in radiology, including digitization, new display and input devices, and the rapid developments of various imaging modalities. Additional radiologic demands, including intelligent support and workflow-specific PACS, functional and quantitative imaging, and CAD and organ-specific workstations, may give rise to several new megatrends. Finally, robotic interventions for biopsy and theragnosis, as well as for imaging genomics, are likely to set off another such wave in the near future. These innovations will expand the role of medical imaging in both diagnosis and treatment.
Go to : 
References
1. Bell ME. Science and art in Roentgenography. Xray Tech. 1948; 20:146–148.
3. DICOM. http://medical.nema.org/Dicom/.
4. IHE. http://ihe.net.
5. HL7. http://www.hl7.org.
6. Chen J, Bradshaw J, Nagy P. Has the Picture Archiving and Communication System (PACS) become a commodity? J Digit Imaging. 2011; 24:6–10. PMID: 20419387.

7. Bandon D, Lovis C, Geissbühler A, Vallée JP. Enterprise-wide PACS: beyond radiology, an architecture to manage all medical images. Acad Radiol. 2005; 12:1000–1009. PMID: 16087095.
8. Law MY, Liu B, Chan LW. Informatics in radiology: DICOM-RT-based electronic patient record information system for radiation therapy. Radiographics. 2009; 29:961–972. PMID: 19448106.
9. Law MY, Liu B. Informatics in radiology: DICOM-RT and its utilization in radiation therapy. Radiographics. 2009; 29:655–667. PMID: 19270073.
10. Zhou Z, Liu BJ, Le AH. CAD-PACS integration tool kit based on DICOM secondary capture, structured report and IHE workflow profiles. Comput Med Imaging Graph. 2007; 31:346–352. PMID: 17386997.

11. Samei E, Seibert JA, Andriole K, Badano A, Crawford J, Reiner B, et al. AAPM/RSNA tutorial on equipment selection: PACS equipment overview: general guidelines for purchasing and acceptance testing of PACS equipment. Radiographics. 2004; 24:313–334. PMID: 14730055.
12. Meyers PH, Nice CM Jr, Becker HC, Nettleton WJ Jr, Sweeney JW, Meckstroth GR. Automated computer analysis of radiographic images. Radiology. 1964; 83:1029–1034. PMID: 14226800.

13. Kruger RP, Townes JR, Hall DL, Dwyer SJ 3rd, Lodwick GS. Automated radiographic diagnosis via feature extraction and classification of cardiac size and shape descriptors. IEEE Trans Biomed Eng. 1972; 19:174–186. PMID: 4553674.

14. Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph. 2007; 31:198–211. PMID: 17349778.

15. Loo LN, Doi K, Metz CE. Investigation of basic imaging properties in digital radiography. 4. Effect of unsharp masking on the detectability of simple patterns. Med Phys. 1985; 12:209–214. PMID: 4000078.

16. Giger ML, Doi K. Investigation of basic imaging properties in digital radiography. 3. Effect of pixel size on SNR and threshold contrast. Med Phys. 1985; 12:201–208. PMID: 4000077.

17. Giger ML, Doi K. Investigation of basic imaging properties in digital radiography. I. Modulation transfer function. Med Phys. 1984; 11:287–295. PMID: 6738452.

18. Freer TW, Ulissey MJ. Screening mammography with computer-aided detection: prospective study of 12,860 patients in a community breast center. Radiology. 2001; 220:781–786. PMID: 11526282.

19. Birdwell RL, Bandodkar P, Ikeda DM. Computer-aided detection with screening mammography in a university hospital setting. Radiology. 2005; 236:451–457. PMID: 16040901.

20. Cupples TE, Cunningham JE, Reynolds JC. Impact of computer-aided detection in a regional screening mammography program. AJR Am J Roentgenol. 2005; 185:944–950. PMID: 16177413.

21. Morton MJ, Whaley DH, Brandt KR, Amrami KK. Screening mammograms: interpretation with computer-aided detection--prospective evaluation. Radiology. 2006; 239:375–383. PMID: 16569779.

22. Butler SA, Gabbay RJ, Kass DA, Siedler DE, O'shaughnessy KF, Castellino RA. Computer-aided detection in diagnostic mammography: detection of clinically unsuspected cancers. AJR Am J Roentgenol. 2004; 183:1511–1515. PMID: 15505329.

23. Warren Burhenne LJ, Wood SA, D'Orsi CJ, Feig SA, Kopans DB, O'Shaughnessy KF, et al. Potential contribution of computer-aided detection to the sensitivity of screening mammography. Radiology. 2000; 215:554–562. PMID: 10796939.

24. Ozekes S, Osman O, Camurcu AY. Mammographic mass detection using a mass template. Korean J Radiol. 2005; 6:221–228. PMID: 16374079.

25. Li F, Arimura H, Suzuki K, Shiraishi J, Li Q, Abe H, et al. Computer-aided detection of peripheral lung cancers missed at CT: ROC analyses without and with localization. Radiology. 2005; 237:684–690. PMID: 16244277.

26. Giger ML, Doi K, MacMahon H. Image feature analysis and computer-aided diagnosis in digital radiography. 3. Automated detection of nodules in peripheral lung fields. Med Phys. 1988; 15:158–166. PMID: 3386584.

27. Song KD, Chung MJ, Kim HC, Jeong SY, Lee KS. Usefulness of the CAD system for detecting pulmonary nodule in real clinical practice. Korean J Radiol. 2011; 12:163–168. PMID: 21430932.

28. Goo JM. A computer-aided diagnosis for evaluating lung nodules on chest CT: the current status and perspective. Korean J Radiol. 2011; 12:145–155. PMID: 21430930.

29. Ozekes S, Osman O, Ucan ON. Nodule detection in a lung region that's segmented with using genetic cellular neural networks and 3D template matching with fuzzy rule based thresholding. Korean J Radiol. 2008; 9:1–9. PMID: 18253070.

30. Kasai S, Li F, Shiraishi J, Li Q, Doi K. Computerized detection of vertebral compression fractures on lateral chest radiographs: preliminary results with a tool for early detection of osteoporosis. Med Phys. 2006; 33:4664–4674. PMID: 17278819.

31. Liberman UA, Weiss SR, Bröll J, Minne HW, Quan H, Bell NH, et al. The Alendronate Phase III Osteoporosis Treatment Study Group. Effect of oral alendronate on bone mineral density and the incidence of fractures in postmenopausal osteoporosis. N Engl J Med. 1995; 333:1437–1443. PMID: 7477143.

32. Hirai T, Korogi Y, Arimura H, Katsuragawa S, Kitajima M, Yamura M, et al. Intracranial aneurysms at MR angiography: effect of computer-aided diagnosis on radiologists' detection performance. Radiology. 2005; 237:605–610. PMID: 16179404.

33. Wardlaw JM, White PM. The detection and management of unruptured intracranial aneurysms. Brain. 2000; 123(Pt 2):205–221. PMID: 10648430.

34. Shiraishi J, Li Q, Appelbaum D, Pu Y, Doi K. Development of a computer-aided diagnostic scheme for detection of interval changes in successive whole-body bone scans. Med Phys. 2007; 34:25–36. PMID: 17278486.

35. Bogoni L, Ko JP, Alpert J, Anand V, Fantauzzi J, Florin CH, et al. Impact of a computer-aided detection (CAD) system integrated into a picture archiving and communication system (PACS) on reader sensitivity and efficiency for the detection of lung nodules in thoracic CT exams. J Digit Imaging. 2012; 25:771–781. PMID: 22710985.

36. Welter P, Hocken C, Deserno TM, Grouls C, Günther RW. Workflow management of content-based image retrieval for CAD support in PACS environments based on IHE. Int J Comput Assist Radiol Surg. 2010; 5:393–400. PMID: 20379792.

37. Le AH, Liu B, Huang HK. Integration of computer-aided diagnosis/detection (CAD) results in a PACS environment using CAD-PACS toolkit and DICOM SR. Int J Comput Assist Radiol Surg. 2009; 4:317–329. PMID: 20033579.

38. Zhou Z. Data security assurance in CAD-PACS integration. Comput Med Imaging Graph. 2007; 31:353–360. PMID: 17387002.

39. Cho HC, Hadjiiski L, Sahiner B, Chan HP, Helvie M, Paramagul C, et al. Similarity evaluation in a content-based image retrieval (CBIR) CADx system for characterization of breast masses on ultrasound images. Med Phys. 2011; 38:1820–1831. PMID: 21626916.

40. Baldi A, Murace R, Dragonetti E, Manganaro M, Guerra O, Bizzi S, et al. Definition of an automated Content-Based Image Retrieval (CBIR) system for the comparison of dermoscopic images of pigmented skin lesions. Biomed Eng Online. 2009; 8:18. PMID: 19682395.

41. Xue Z, Antani S, Long LR, Jeronimo J, Thoma GR. Investigating CBIR techniques for cervicographic images. AMIA Annu Symp Proc. 2007; 826–830. PMID: 18693952.
42. Kherfi ML, Ziou D. Relevance feedback for CBIR: a new approach based on probabilistic feature weighting with positive and negative examples. IEEE Trans Image Process. 2006; 15:1017–1030. PMID: 16579386.

43. Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999; 9:195–207. PMID: 9931269.
44. Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999; 9:179–194. PMID: 9931268.
45. Birmingham University Imaging Centre. Using the Brain Imaging Lab (BIL). Accessed November 18, 2011. http://www.buic.bham.ac.uk/pbic/bilusage.html.
46. Wang Y, Gupta A, Liu Z, Zhang H, Escolar ML, Gilmore JH, et al. DTI registration in atlas based fiber analysis of infantile Krabbe disease. Neuroimage. 2011; 55:1577–1586. PMID: 21256236.

47. Lee CW, Seo JB, Lee Y, Chae EJ, Kim N, Lee HJ, et al. A pilot trial on pulmonary emphysema quantification and perfusion mapping in a single-step using contrast-enhanced dual-energy computed tomography. Invest Radiol. 2012; 47:92–97. PMID: 21750465.
48. Park SO, Seo JB, Kim N, Lee YK, Lee J, Kim DS. Comparison of usual interstitial pneumonia and nonspecific interstitial pneumonia: quantification of disease severity and discrimination between two diseases on HRCT using a texture-based automated system. Korean J Radiol. 2011; 12:297–307. PMID: 21603289.

49. Lim J, Kim N, Seo JB, Lee YK, Lee Y, Kang SH. Regional context-sensitive support vector machine classifier to improve automated identification of regional patterns of diffuse interstitial lung disease. J Digit Imaging. 2011; 24:1133–1140. PMID: 21311944.

50. Lee HJ, Seo JB, Chae EJ, Kim N, Lee CW, Oh YM, et al. Tracheal morphology and collapse in COPD: correlation with CT indices and pulmonary function test. Eur J Radiol. 2011; 80:e531–e535. PMID: 21315531.

51. Chae EJ, Kim TB, Cho YS, Park CS, Seo JB, Kim N, et al. Airway Measurement for Airway Remodeling Defined by Post-Bronchodilator FEV1/FVC in Asthma: Investigation Using Inspiration-Expiration Computed Tomography. Allergy Asthma Immunol Res. 2011; 3:111–117. PMID: 21461250.

52. Chae EJ, Seo JB, Song JW, Kim N, Park BW, Lee YK, et al. Slope of emphysema index: an objective descriptor of regional heterogeneity of emphysema and an independent determinant of pulmonary function. AJR Am J Roentgenol. 2010; 194:W248–W255. PMID: 20173123.

53. Park SO, Seo JB, Kim N, Park SH, Lee YK, Park BW, et al. Feasibility of automated quantification of regional disease patterns depicted on high-resolution computed tomography in patients with various diffuse lung diseases. Korean J Radiol. 2009; 10:455–463. PMID: 19721830.

54. Kim N, Seo JB, Song KS, Chae EJ, Kang SH. Semi-automatic measurement of the airway dimension by computed tomography using the full-with-half-maximum method: a study of the measurement accuracy according to the orientation of an artificial airway. Korean J Radiol. 2008; 9:236–242. PMID: 18525226.

55. Kim N, Seo JB, Song KS, Chae EJ, Kang SH. Semi-automatic measurement of the airway dimension by computed tomography using the full-width-half-maximum method: a study on the measurement accuracy according to the CT parameters and size of the airway. Korean J Radiol. 2008; 9:226–235. PMID: 18525225.

56. Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci U S A. 1990; 87:9868–9872. PMID: 2124706.

57. Whalley HC, Papmeyer M, Sprooten E, Lawrie SM, Sussmann JE, McIntosh AM. Review of functional magnetic resonance imaging studies comparing bipolar disorder and schizophrenia. Bipolar Disord. 2012; 14:411–431. PMID: 22631622.

58. Astrakas LG, Naqvi SH, Kateb B, Tzika AA. Functional MRI using robotic MRI compatible devices for monitoring rehabilitation from chronic stroke in the molecular medicine era (Review). Int J Mol Med. 2012; 29:963–973. PMID: 22426741.

59. Paloyelis Y, Mehta MA, Kuntsi J, Asherson P. Functional MRI in ADHD: a systematic literature review. Expert Rev Neurother. 2007; 7:1337–1356. PMID: 17939771.

60. Crosson B, McGregor K, Gopinath KS, Conway TW, Benjamin M, Chang YL, et al. Functional MRI of language in aphasia: a review of the literature and the methodological challenges. Neuropsychol Rev. 2007; 17:157–177. PMID: 17525865.

61. Powell HW, Koepp MJ, Richardson MP, Symms MR, Thompson PJ, Duncan JS. The application of functional MRI of memory in temporal lobe epilepsy: a clinical review. Epilepsia. 2004; 45:855–863. PMID: 15230713.

62. Histed SN, Lindenberg ML, Mena E, Turkbey B, Choyke PL, Kurdziel KA. Review of functional/anatomical imaging in oncology. Nucl Med Commun. 2012; 33:349–361. PMID: 22314804.

63. Del Vecchio S, Zannetti A, Fonti R, Iommelli F, Pizzuti LM, Lettieri A, et al. PET/CT in cancer research: from preclinical to clinical applications. Contrast Media Mol Imaging. 2010; 5:190–200. PMID: 20812287.

64. Cai W, Chen X. Multimodality molecular imaging of tumor angiogenesis. J Nucl Med. 2008; 49(Suppl 2):113S–128S. PMID: 18523069.

65. Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC. Group. A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology. 2011; 258:906–914. PMID: 21339352.

66. Buckler AJ, Mulshine JL, Gottlieb R, Zhao B, Mozley PD, Schwartz L. The use of volumetric CT as an imaging biomarker in lung cancer. Acad Radiol. 2010; 17:100–106. PMID: 19969253.

67. Buckler AJ, Mozley PD, Schwartz L, Petrick N, McNitt-Gray M, Fenimore C, et al. Volumetric CT in lung cancer: an example for the qualification of imaging as a biomarker. Acad Radiol. 2010; 17:107–115. PMID: 19969254.
68. Frank R. FDG-PET/CT Working Group. Quantitative Imaging Biomarkers Alliance FDG-PET/CT Working Group report. Mol Imaging Biol. 2008; 10:305. PMID: 18704590.

69. Smith JJ, Sorensen AG, Thrall JH. Biomarkers in imaging: realizing radiology's future. Radiology. 2003; 227:633–638. PMID: 12663828.

70. Clarke LP, Sriram RD, Schilling LB. Imaging as a Biomarker: Standards for Change Measurements in Therapy workshop summary. Acad Radiol. 2008; 15:501–530. PMID: 18389935.
71. McLennan G, Clarke L, Hohl RJ. Imaging as a biomarker for therapy response: cancer as a prototype for the creation of research resources. Clin Pharmacol Ther. 2008; 84:433–436. PMID: 18802422.

72. Armato SG 3rd, Meyer CR, Mcnitt-Gray MF, McLennan G, Reeves AP, Croft BY, et al. The Reference Image Database to Evaluate Response to therapy in lung cancer (RIDER) project: a resource for the development of change-analysis software. Clin Pharmacol Ther. 2008; 84:448–456. PMID: 18754000.

73. Petrick N, Brown DG, Suleiman O, Myers KJ. Imaging as a tumor biomarker in oncology drug trials for lung cancer: the FDA perspective. Clin Pharmacol Ther. 2008; 84:523–525. PMID: 18716616.

74. Gavrielides MA, Kinnard LM, Myers KJ, Petrick N. Noncalcified lung nodules: volumetric assessment with thoracic CT. Radiology. 2009; 251:26–37. PMID: 19332844.

75. Quantitative Imaging Biomarkers Alliance. http://www.rsna.org/QIBA_.aspx. 2009.
76. Goo HW, Yang DH, Kim N, Park SI, Kim DK, Kim EA. Collateral ventilation to congenital hyperlucent lung lesions assessed on xenon-enhanced dynamic dual-energy CT: an initial experience. Korean J Radiol. 2011; 12:25–33. PMID: 21228937.

77. Chae EJ, Seo JB, Lee J, Kim N, Goo HW, Lee HJ, et al. Xenon ventilation imaging using dual-energy computed tomography in asthmatics: initial experience. Invest Radiol. 2010; 45:354–361. PMID: 20404734.
78. NVIDIA 3D Vision. http://www.nvidia.com/object/3d-vision-main.html.
79. Okoshi T. Three Dimensional Imaging Techniques. 1976. New York: Academic Press.
80. Sandin D, Margolis T, Dawe G, Leigh J, DeFanti T. The Varrier Autostereographic Display. Proc SPIE. 2001; 4297:204–211.
81. Holographics North. What's New. http://www.holonorth.com/new.htm.
82. Soares OD, Fernandes JC. Cylindrical hologram of 360 degrees field of view. Appl Opt. 1982; 21:3194–3196. PMID: 20396202.
83. Kutter O, Aichert A, Bichlmeier C, Traub J, Heining SM, Ockert B, et al. Real-time volume rendering for high quality visualization in augmented reality. International Workshop on Augmented environments for Medical Imaging including Augmented Reality in Computer-aided Surgery (AMI-ARCS 2008). 2008. New York.
84. Werkgartner G, Lemmerer M, Hauser H, Sorantin E, Beichel R, Reitinger B, et al. Augmented-reality-based liver-surgical planning system. Eur Surg. 2004; 36:270–274.

87. Glahn DC, Paus T, Thompson PM. Imaging genomics: mapping the influence of genetics on brain structure and function. Hum Brain Mapp. 2007; 28:461–463. PMID: 17471577.

88. Thompson PM, Martin NG, Wright MJ. Imaging genomics. Curr Opin Neurol. 2010; 23:368–373. PMID: 20581684.

89. Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer's disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008; 43:458–469. PMID: 18691658.

90. Pievani M, Rasser PE, Galluzzi S, Benussi L, Ghidoni R, Sabattoli F, et al. Mapping the effect of APOE epsilon4 on gray matter loss in Alzheimer's disease in vivo. Neuroimage. 2009; 45:1090–1098. PMID: 19349226.
91. Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, et al. MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009; 132(Pt 4):1067–1077. PMID: 19251758.

92. Filippini N, Zarei M, Beckmann CF, Galluzzi S, Borsci G, Testa C, et al. Regional atrophy of transcallosal prefrontal connections in cognitively normal APOE epsilon4 carriers. J Magn Reson Imaging. 2009; 29:1021–1026. PMID: 19388128.
93. Shaw P, Lerch JP, Pruessner JC, Taylor KN, Rose AB, Greenstein D, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 2007; 6:494–500. PMID: 17509484.

94. van Nuenen BF, van Eimeren T, van der Vegt JP, Buhmann C, Klein C, Bloem BR, et al. Mapping preclinical compensation in Parkinson's disease: an imaging genomics approach. Mov Disord. 2009; 24(Suppl 2):S703–S710. PMID: 19877238.

95. Guccione S, Yang YS, Shi G, Lee DY, Li KC, Bednarski MD. Functional genomics guided with MR imaging: mouse tumor model study. Radiology. 2003; 228:560–568. PMID: 12821773.

96. Borgwardt SJ, Fusar-Poli P. Imaging Genomics - An integrative approach to understand the biological susceptibility for schizophrenia. Med Hypotheses. 2007; 68:1426. PMID: 17161548.

97. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. PMID: 12045153.

98. Goecks J, Nekrutenko A, Taylor J. Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010; 11:R86. PMID: 20738864.

99. Nicholas BL, O'Connor CD, Djukanovic R. From proteomics to prescription-the search for COPD biomarkers. COPD. 2009; 6:298–303. PMID: 19811390.

100. Fava C, Montagnana M, Guidi GC, Melander O. From circulating biomarkers to genomics and imaging in the prediction of cardiovascular events in the general population. Ann Med. 2012; 44:433–447. PMID: 21623699.

101. Chen SS, Wang YP. Translational systems genomics: ontology and imaging. Summit on Translat Bioinforma. 2009; 2009:21–25. PMID: 21347165.
102. Čapek K, Novack C. R.U.R. (Rossum's Universal Robots). 2004. New York: Penguin Books.
103. Hockstein N, Gourin C, Faust R, Terris D. A history of robots: from science fiction to surgical robotics. J Robot Surg. 2007; 1:113–118.

104. Bargar WL, Bauer A, Börner M. Primary and revision total hip replacement using the Robodoc system. Clin Orthop Relat Res. 1998; 82–91. PMID: 9755767.

105. Gourin CG, Terris DJ. Surgical robotics in otolaryngology: expanding the technology envelope. Curr Opin Otolaryngol Head Neck Surg. 2004; 12:204–208. PMID: 15167030.

106. Ballantyne GH. Robotic surgery, telerobotic surgery, telepresence, and telementoring. Review of early clinical results. Surg Endosc. 2002; 16:1389–1402. PMID: 12140630.
107. Himpens J, Leman G, Cadiere GB. Telesurgical laparoscopic cholecystectomy. Surg Endosc. 1998; 12:1091. PMID: 9685550.

108. Rockall TA. Ballantyne GH, Marescaux J, Giulianotti PC, editors. The da Vinci telerobotic surgical system. Primer of robotic & telerobotic surgery. 2004. Philadelphia: Lippincott Williams & Wilkins;p. 57–60.
109. Neubach Z, Shoham M. Ultrasound-guided robot for flexible needle steering. IEEE Trans Biomed Eng. 2010; 57:799–805. PMID: 19709957.

110. Chun KR, Schmidt B, Köktürk B, Tilz R, Fürnkranz A, Konstantinidou M, et al. Catheter ablation - new developments in robotics. Herz. 2008; 33:586–589. PMID: 19137249.

111. Ernst S. Robotic approach to catheter ablation. Curr Opin Cardiol. 2008; 23:28–31. PMID: 18281824.

112. Marcelli E, Cercenelli L, Plicchi G. A novel telerobotic system to remotely navigate standard electrophysiology catheters. Comput Cardiol. 2008; 14-17:137–140.

113. Reddy VY, Neuzil P, Malchano ZJ, Vijaykumar R, Cury R, Abbara S, et al. View-synchronized robotic image-guided therapy for atrial fibrillation ablation: experimental validation and clinical feasibility. Circulation. 2007; 115:2705–2714. PMID: 17502570.
Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download