Abstract
Human organic cation/carnitine transporter 1 (OCTN1) plays an important role in the transport of drugs and endogenous substances. It is known that a missense variant of OCTN1 is significantly associated with Crohn's disease susceptibility. This study was performed to identify genetic variants of the OCTN1 promoter in Korean individuals and to determine their functional effects. First, the promoter region of OCTN1 was directly sequenced using genomic DNA samples from 48 healthy Koreans. OCTN1 promoter activity was then measured using a luciferase reporter assay in HCT-116 cells. Seven variants of the OCTN1 promoter were identified, two of which were novel. There were also four major OCTN1 promoter haplotypes. Three haplotypes (H1, H3, and H4) showed decreased transcriptional activity, which was reduced by 22.9%, 23.0%, and 44.6%, respectively (p<0.001), compared with the reference haplotype (H2). Transcription factor binding site analyses and gel shift assays revealed that NF-Y could bind to the region containing g.-1875T>A, a variant present in H3, and that the binding affinity of NF-Y was higher for the g.-1875T allele than for the g.-1875A allele. NF-Y could also repress OCTN1 transcription. These data suggest that three OCTN1 promoter haplotypes could regulate OCTN1 transcription. To our knowledge, this is the first study to identify functional variants of the OCTN1 promoter.
Human organic cation/carnitine transporter 1 (OCTN1, SLC22A4) belongs to the family of solute carrier (SLC) transporters, and is expressed mainly in the kidney, skeletal muscle, trachea, bone marrow, and fetal liver [1,2]. OCTN1 is also expressed in the small intestine, liver, brain neurons, and inflammatory joints in mice [3,4,5]. It is widely accepted that OCTN1 plays an important role in transporting organic cations, including tetraethylammonium (TEA), in a pH-dependent manner [2,6]. Recently, Gründemann et al. [7] reported that L-ergothioneine is a natural substrate of the OCTN1 transporter.
Many previous studies reported that a missense variant of OCTN1 is associated with the susceptibility to, or progression of, human diseases, particularly Crohn's disease [8,9,10,11,12,13,14,15]. For example, Peltekova et al. [8] demonstrated that L503F of OCTN1 showed increased transport activity for TEA while it had decreased transport activity for other endogenous compounds including carnitine through in vitro uptake assay. In addition, they found that the haplotype consisting of L503F of OCTN1 and g.-207G>C of OCTN2 was significantly associated with susceptibility to Crohn's disease. This finding was validated in subsequent studies [9,10,11,12,13,14].
Although there are few known substrates of OCTN1, genetic variants of this transporter could alter the pharmacokinetics or pharmacodynamics of drugs that are OCTN1 substrates. L503F was previously shown to have decreased transport activity for the anticonvulsant drug gabapentin, and this variant resulted in significantly decreased renal secretion of gabapentin [16]. In another example, Nakamichi et al. [17] reported that metformin is an OCTN1 substrate, and that its maximum plasma concentration (Cmax) could be changed significantly in OCTN1 knockout mice.
To date, there have been few studies investigating the effect of genetic variations of OCTN1 promoter. Recently, 272 DNA samples from ethnically diverse populations were directly sequenced; six OCTN1 promoter variants were identified and eight haplotypes were functionally characterized [18]. However, no haplotype exhibited significantly altered promoter activity compared with the reference haplotype in that study. Maeda et al. [19] investigated the regulatory mechanisms of OCTN1 expression, and found that it could be regulated by several transcription factors including RUNX1, Sp1, and NF-κB.
The present study was performed to functionally characterize genetic variations in the OCTN1 promoter region in Korean individuals, and to determine the mechanism by which OCTN1 variants alter promoter activity. First, we performed genetic analysis using 48 genomic DNA samples from healthy Korean individuals to identify variants of the OCTN1 promoter. We then constructed the common OCTN1 promoter haplotypes, and investigated the function of each haplotype using in vitro assays including dual-luciferase reporter and electrophoretic mobility shift assays. In this study, novel promoter variants of OCTN1 in Korean individuals were identified, and the mechanism responsible for the transcriptional regulation of this gene was determined.
This study was approved by the Institutional Review Board of Ewha Medical Center, Seoul, Republic of Korea (ECT 12-15-02). Forty-eight genomic DNA samples were collected from healthy Korean individuals from the DNA bank of the Korea Pharmacogenomics Research Network at Seoul National University, Seoul, Republic of Korea. All subjects enrolled in this study had an East Asian ethnic background. To identify genetic variants in the promoter region of OCTN1, a 2,345 base pair (bp) polymerase chain reaction (PCR) fragment (-2,240 to -105 bp from the translation start site) was amplified and directly sequenced using an automated genetic analyzer (Life Technologies, Carlsbad, CA). Haplotype assembly was performed using the Haploview program (version 4.2, developed by the Broad Institute, Cambridge, MA). Nucleotide location numbers were assigned from the translational start site based on the OCTN1 mRNA sequence (GenBank accession number; NM_003059.2).
To construct a reporter plasmid containing a promoter region with the OCTN1 reference sequence, a 2,127 bp of the OCTN1 gene was amplified using primers containing recognition sites for the restriction endonucleases XhoI and HindIII (Table 1) from genomic DNA sample from the individual with a reference sequence according to NM_003059.2. The amplified product was then inserted into the pGL4.11b [luc2] vector. Genetic variants in the promoter region were introduced into the pGL4.11b-OCTN1 vector using a QuikChange® II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) using the primers listed in Table 1. The DNA sequences were confirmed by direct sequencing.
Reporter assays were performed as described previously [17]. First, reporter plasmids containing either the reference or variant sequences of OCTN1 were transfected into HCT-116 (human colon carcinoma) cells using Lipofectamine LTX and Plus reagents (Life Technologies). Thirty hours after transfection, reporter activities were measured and quantified using a Dual-luciferase® reporter assay system and a Glomax 96-well plate luminometer (Promega). Relative luciferase activity was defined as the ratio of firefly luciferase to renilla luciferase. To examine the effect of nuclear factor-Y (NF-Y) on OCTN1 promoter activity, reference or variant reporter plasmids were co-transfected with increasing amounts (5~20 ng) of NF-Y-pcDNA3.1(+) vector. The NF-Y-pcDNA3.1(+) vector was constructed previously [20].
Electrophoretic mobility shift assays (EMSAs) were performed as described previously [20]. Nuclear protein extracts were obtained from HCT-116 cells, and 20 or 35µg of the extracts were incubated with a 32P-labeled oligonucleotide (2×105 counts/min) for 30 min at room temperature. For the competition assay, unlabeled NF-Y consensus or mutant oligonucleotides were added into the nuclear extracts prior to the binding reaction. For the supershift assay, 4.2µl of a mixture of NF-YA (sc-7712, Santa Cruz Biotechnology, Santa Cruz, CA), NF-YB (sc-7711X, Santa Cruz Biotechnology), and NF-YC (sc-7714X, Santa Cruz Biotechnology) antibodies or 2µl of a myeloid zinc finger-1 (MZF-1) antibody (sc-46178X, Santa Cruz Biotechnology) were incubated with nuclear extracts for 30 min at room temperature prior to the binding reaction. Table 1 lists the oligonucleotides that were used in the EMSAs. The reaction mixtures were loaded onto a 6% non-denaturing polyacrylamide gel, and electrophoresed for 70 min at 80 V. The gel was dried, and then exposed to CP-BU film (Agfa, Mortsel, Belgium) for 16 h at -80℃. The intensity of each band was measured using ImageJ software (National Institutes of Health, Bethesda, MD).
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA). p values for the luciferase assay were calculated using one-way analysis of variance followed by Dunnett's two-tailed test. A paired t-test was used to compare the effects of NF-Y on the reference and g.-1875T>A variant OCTN1 promoters. p<0.05 was considered to be statistically significant.
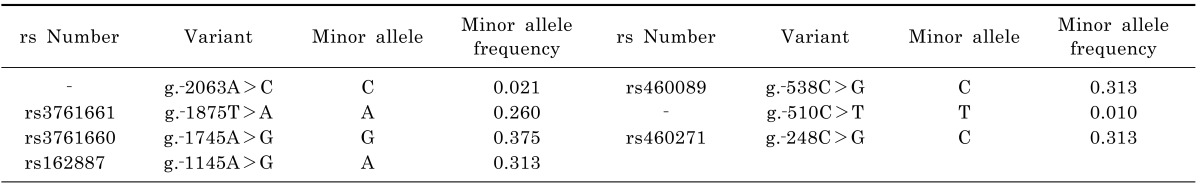
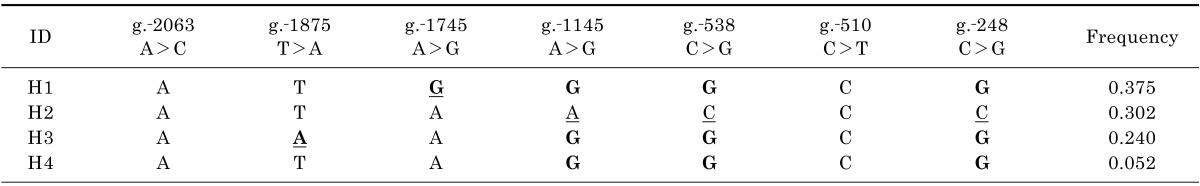
In this study, seven variants of the OCTN1 promoter region were identified, and two of these were novel (Table 2). The name of each variant follows the Human Genome Variation Society (HGVS) nomenclature. Table 3 lists the frequencies of the common (frequency ≥5%) haplotypes, which were assembled using the individual genetic data. Haplotype 1 (H1), consisting of all the major alleles except for one minor allele at g.-1745A>G, was the most common haplotype in the study population. Haplotype 2 (H2) includes three minor alleles at g.-1145A>G, g.-538C>G, and g.-248C>G, and contains a reference sequence according to NM_003059.2. Therefore, this haplotype was used as the OCTN1 reference haplotype in the present study.
To date, no studies have investigated the function of genetic variants of the OCTN1 promoter using in vitro assays, although the function of proximal promoter haplotypes of OCTN1 has been reported [18]. Therefore, to examine the effects of the variants on OCTN1 promoter activity, we constructed the reporter plasmids containing either reference or variants of OCTN1, and performed luciferase assays after transfecting these plasmids into HCT-116 cells. Previously four cell lines, ACHN (human kidney adenocarcinoma), HepG2 (human liver carcinoma), HeLa (human uterus carcinoma), and HCT-116 cells were tested for measuring OCTN1 promoter activity, and among them the OCTN1-reference-HCT-116 cells showed the highest promoter activity [18]. Therefore, HCT-116 cells were used to perform the luciferase assays in this study. Fig. 1a shows the relative luciferase activities of the OCTN1 promoter haplotypes. All three haplotypes containing the single nucleotide polymorphisms (SNPs) showed significantly decreased promoter activities, which were reduced by 23~45% compared with the reference. We next measured the luciferase activity of each SNP present in the haplotypes. Two of the five variants (g.-1875T>A and g.-1745A>G) showed increased promoter activities, whereas two (g.-1145A>G and g.-248C>G) had decreased promoter activities; the activity of g.-538C>G was comparable with the reference (Fig. 1b). These findings were consistent with the luciferase assay results obtained for the OCTN1 haplotypes: the activities of H1 and H3 were higher than H4. All three haplotypes that exhibited decreased promoter activities contained g.-1145A>G and g.-248C>G. However, H1 and H3 also had the variants g.-1745A>G and g.-1875T>A, respectively, which showed increased promoter activities.
To further investigate the mechanisms of transcriptional regulation of OCTN1 promoter variants, we used TFSearch (version 1.3, developed by RWCP, Tokyo, Japan) and MatInspector (Genomatix Software GmbH, Munich, Germany) to identify transcription factors that could bind to the promoter region of OCTN1 in the vicinity of each variant present in haplotypes 1, 3, or 4. The results predicted that only one variant, g.-1875T>A, might affect the binding affinity of the transcriptional factor NF-Y. In other words, transcription factor binding site (TFBS) analyses predicted that NF-Y would have a higher binding affinity for the reference g.-1875T allele than for the variant g.-1875A allele.
To validate this prediction, we performed EMSAs, in which labeled oligonucleotides (2×105 counts/minute, NF-Y consensus: lanes 1~3; reference, g.-1875T: lanes 4~6; variant, g.-1875A: lanes 7~9, Fig. 2a) were incubated with nuclear protein extracts. Competition assay were also performed by incubating the nuclear extracts with a 50-fold concentration of unlabeled NF-Y consensus (lanes 2, 5, and 8) or mutant (lanes 3, 6, and 9) oligonucleotides prior to the binding reaction (Fig. 2a). The reference probe formed DNA-protein complexes that were detected in the same location as the NF-Y consensus probe, but with a decreased intensity (lanes 1 and 4, Fig. 2a). However, the intensity of the DNA-protein complexes with the variant probe was very weak (lane 7, Fig. 2a). In the competition assay, unlabeled NF-Y consensus oligonucleotides competed with the NF-Y or reference probes (lanes 2 and 5, Fig. 2a). However, these binding complexes were unable to compete with unlabeled oligonucleotides containing mutated core NF-Y sequences (lanes 3 and 6, Fig. 2a). We also performed a supershift assay using a mixture of NF-YA, NF-YB, and NF-YC antibodies. A supershift in the presence of antibodies against NF-Y confirmed that NF-Y was present in the complex (lanes 2, 4, and 5, Fig. 2b). To confirm the results of the supershift assay, we performed an additional assay using a non-NF-Y-specific antibody against MZF-1, which did not induce a supershift (lane 3, Fig. 2b).
The effect of NF-Y on OCTN1 promoter activity was investigated by overexpressing NF-Y and subsequently conducting luciferase assays. NF-Y overexpression resulted in a dose-dependent reduction in OCTN1 promoter activity. NF-Y exerted stronger effects on promoter activity in the presence of the reference g.-1875T than the variant g.-1875A (Fig. 3). These findings are consistent with the luciferase assay results obtained using OCTN1 variants, where g.-1875T>A showed significantly increased promoter activity compared with the reference.
This study was performed to identify and functionally characterize genetic variations in the OCTN1 promoter region in Korean individuals. Direct sequencing identified seven variants, of which six were polymorphic (Table 2). Previously, Tahara et al. [18] performed genetic analysis of the OCTN1 proximal promoter by screening 272 DNA samples from 68 Caucasian, 68 Chinese, 68 Hispanic, and 68 African individuals. They identified seven genetic variants of the OCTN1 promoter, and only two of these variants, g.-256C>G and g.-248C>G, were found in Chinese individuals. The minor allele frequency of g.-248C>G in the Tahara et al. study was similar to that in the current study. However, the rare variant g.-256C>G, which had a frequency of 1.5% in the Tahara et al. study, was not observed in our Korean population. In the current study, we screened a wide range of the promoter region of OCTN1 (up to -2,240 bp from the translational start site), and identified two novel variants, g.-2063A>C and g.-510C>T. The region from -104 bp to -1 bp upstream of the OCTN1 translational start site was not included in our study's genetic analysis owing to technical problems. Previously, the frequency of a known OCTN1 promoter SNP within -105 bp of the start site, rs11568501 was examined by genotyping DNA samples from 90 healthy Korean individuals [21]. In that population, this SNP was not detected. In addition, Tahara et al. [18] also were unable to detect any SNP within -105 bp of the OCTN1 translational start site. Therefore, we expected to find no SNP within the specified range in our population. Furthermore, the frequencies of OCTN1 promoter haplotypes containing common SNPs in the present study is not affected by performing an additional genetic analysis of OCTN1, in which the more proximal promoter region of this gene is included.
Luciferase reporter assays revealed that three of the OCTN1 haplotypes (H1, H3, and H4) had significantly decreased promoter activity (Fig. 1a). These three haplotypes contained the variants g.-1145A>G, g.-538C>G, and g.-248C>G. Of these variants, g.-1145A>G and g.-248C>G showed significantly decreased promoter activities, while the promoter activity of g.-538C>G was similar to that of the reference. The promoter activities of H1 and H3 were higher than that of H4, possibly because of the presence of g.-1745A>G and g.-1875T>A, respectively, which showed increased promoter activities. In a previous study, the promoter activities of seven OCTN1 promoter haplotypes containing a combination of several variants of the OCTN1 proximal promoter were measured [18]. However, no haplotypes exhibited significantly different promoter activities in that study.
TFBS analyses suggested that NF-Y could bind to the OCTN1 promoter region containing g.-1875T>A, a variant present in haplotype 3, and that the binding affinity of NF-Y would be variant-dependent. Specifically, our data predicted that NF-Y would have a higher binding affinity for the reference g.-1875T allele than for the variant g.-1875A allele. Previously, we reported that NF-Y is a transcriptional factor involved in regulating multidrug resistance 3 (MDR3) transcription, and that it could induce the expression of this gene [20].
The consensus DNA sequence of NF-Y is YYRRCCAATCAG [22]. Because the CCAAT motif is a common promoter element, NF-Y plays an important role in the transcriptional regulation of various genes [23,24]. In the current study, we observed that OCTN1 contains a CCAAT motif, with the sequence TCGCCAATAAC. In contrast, the variant g.-1875T>A has the sequence TCGCCAAAAAC, which is a poorer match than the reference. The predictions obtained from TFBS analyses were validated using EMSAs, in which the intensity of the DNA-NF-Y complex was decreased by 70% in the presence of g.-1875T>A (Fig. 2a). In addition, NF-Y repressed OCTN1 transcription, in contrast to its role in MDR3 transcription [20] (Fig. 3). Previous studies reported that NF-Y is a bifunctional transcription factor that can either induce or suppress gene expression [25,26].
Recently, several studies reported that a haplotype consisting of the OCTN1 variant L503F and the OCTN2 variant g.-207G>C was significantly associated with susceptibility to Crohn's disease. Another study reported that an intronic variant of OCTN1 was associated with rheumatoid arthritis [3]. However, the results of association studies between OCTN1 and rheumatoid arthritis remain controversial [27,28].
In conclusion, we identified and characterized genetic variants in the promoter region of OCTN1 in Korean individuals. We identified three haplotypes of OCTN1 with significantly decreased promoter activities. In addition, a mechanism by which genetic variants of OCTN1 regulate promoter activity was determined: g.-1875T>A, a variant present in H3, was associated with significantly increased OCTN1 promoter activity, and this observation was related to the reduced binding of NF-Y, a repressor of OCTN1 transcription, to the variant g.-1875A allele. To our knowledge, this is the first study to identify functional variants of the OCTN1 promoter. Future clinical studies are needed to evaluate the effect of each variant or haplotype of OCTN1 on disease susceptibility and the pharmacokinetics or pharmacodynamics of drugs.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) [2010-0029353]; and a grant of the National Project for Personalized Genomic Medicine, Ministry for Health & Welfare, Republic of Korea [A111218-PG03]. We thank Dr. I. Jang and Dr. J.Y. Cho for giving us a great help to get genomic samples from the DNA bank of Korea Pharmacogenomics Research Network at Seoul National University, Seoul, Republic of Korea.
References
1. Tamai I, Yabuuchi H, Nezu J, Sai Y, Oku A, Shimane M, Tsuji A. Cloning and characterization of a novel human pH-dependent organic cation transporter, OCTN1. FEBS Lett. 1997; 419:107–111. PMID: 9426230.
2. Yabuuchi H, Tamai I, Nezu J, Sakamoto K, Oku A, Shimane M, Sai Y, Tsuji A. Novel membrane transporter OCTN1 mediates multispecific, bidirectional, and pH-dependent transport of organic cations. J Pharmacol Exp Ther. 1999; 289:768–773. PMID: 10215651.
3. Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, Suzuki M, Nagasaki M, Ohtsuki M, Ono M, Furukawa H, Nagashima M, Yoshino S, Mabuchi A, Sekine A, Saito S, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. 2003; 35:341–348. PMID: 14608356.

4. Nakamichi N, Taguchi T, Hosotani H, Wakayama T, Shimizu T, Sugiura T, Iseki S, Kato Y. Functional expression of carnitine/organic cation transporter OCTN1 in mouse brain neurons: possible involvement in neuronal differentiation. Neurochem Int. 2012; 61:1121–1132. PMID: 22944603.

5. Sugiura T, Kato S, Shimizu T, Wakayama T, Nakamichi N, Kubo Y, Iwata D, Suzuki K, Soga T, Asano M, Iseki S, Tamai I, Tsuji A, Kato Y. Functional expression of carnitine/organic cation transporter OCTN1/SLC22A4 in mouse small intestine and liver. Drug Metab Dispos. 2010; 38:1665–1672. PMID: 20601551.

6. Kawasaki Y, Kato Y, Sai Y, Tsuji A. Functional characterization of human organic cation transporter OCTN1 single nucleotide polymorphisms in the Japanese population. J Pharm Sci. 2004; 93:2920–2926. PMID: 15459889.

7. Gründemann D, Harlfinger S, Golz S, Geerts A, Lazar A, Berkels R, Jung N, Rubbert A, Schömig E. Discovery of the ergothioneine transporter. Proc Natl Acad Sci U S A. 2005; 102:5256–5261. PMID: 15795384.
8. Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004; 36:471–475. PMID: 15107849.

9. Noble CL, Nimmo ER, Drummond H, Ho GT, Tenesa A, Smith L, Anderson N, Arnott ID, Satsangi J. The contribution of OCTN1/2 variants within the IBD5 locus to disease susceptibility and severity in Crohn's disease. Gastroenterology. 2005; 129:1854–1864. PMID: 16344054.

10. Russell RK, Drummond HE, Nimmo ER, Anderson NH, Noble CL, Wilson DC, Gillett PM, McGrogan P, Hassan K, Weaver LT, Bisset WM, Mahdi G, Satsangi J. Analysis of the influence of OCTN1/2 variants within the IBD5 locus on disease susceptibility and growth indices in early onset inflammatory bowel disease. Gut. 2006; 55:1114–1123. PMID: 16469794.

11. Martínez A, Martín MC, Mendoza JL, Taxonera C, Díaz-Rubio M, de la Concha EG, Urcelay E. Association of the organic cation transporter OCTN genes with Crohn's disease in the Spanish population. Eur J Hum Genet. 2006; 14:222–226. PMID: 16333318.

12. Leung E, Hong J, Fraser AG, Merriman TR, Vishnu P, Krissansen GW. Polymorphisms in the organic cation transporter genes SLC22A4 and SLC22A5 and Crohn's disease in a New Zealand Caucasian cohort. Immunol Cell Biol. 2006; 84:233–236. PMID: 16519742.
13. Lin Z, Nelson L, Franke A, Poritz L, Li TY, Wu R, Wang Y, MacNeill C, Thomas NJ, Schreiber S, Koltun WA. OCTN1 variant L503F is associated with familial and sporadic inflammatory bowel disease. J Crohns Colitis. 2010; 4:132–138. PMID: 21122496.

14. Xuan C, Zhang BB, Yang T, Deng KF, Li M, Tian RJ. Association between OCTN1/2 gene polymorphisms (1672C-T, 207G-C) and susceptibility of Crohn's disease: a meta-analysis. Int J Colorectal Dis. 2012; 27:11–19. PMID: 21706137.

15. Angelini S, Pantaleo MA, Ravegnini G, Zenesini C, Cavrini G, Nannini M, Fumagalli E, Palassini E, Saponara M, Di Battista M, Casali PG, Hrelia P, Cantelli-Forti G, Biasco G. Polymorphisms in OCTN1 and OCTN2 transporters genes are associated with prolonged time to progression in unresectable gastrointestinal stromal tumours treated with imatinib therapy. Pharmacol Res. 2013; 68:1–6. PMID: 23127916.

16. Urban TJ, Brown C, Castro RA, Shah N, Mercer R, Huang Y, Brett CM, Burchard EG, Giacomini KM. Effects of genetic variation in the novel organic cation transporter, OCTN1, on the renal clearance of gabapentin. Clin Pharmacol Ther. 2008; 83:416–421. PMID: 17609685.

17. Nakamichi N, Shima H, Asano S, Ishimoto T, Sugiura T, Matsubara K, Kusuhara H, Sugiyama Y, Sai Y, Miyamoto K, Tsuji A, Kato Y. Involvement of carnitine/organic cation transporter OCTN1/SLC22A4 in gastrointestinal absorption of metformin. J Pharm Sci. 2013; 102:3407–3417. PMID: 23666872.

18. Tahara H, Yee SW, Urban TJ, Hesselson S, Castro RA, Kawamoto M, Stryke D, Johns SJ, Ferrin TE, Kwok PY, Giacomini KM. Functional genetic variation in the basal promoter of the organic cation/carnitine transporters OCTN1 (SLC22A4) and OCTN2 (SLC22A5). J Pharmacol Exp Ther. 2009; 329:262–271. PMID: 19141711.

19. Maeda T, Hirayama M, Kobayashi D, Miyazawa K, Tamai I. Mechanism of the regulation of organic cation/carnitine transporter 1 (SLC22A4) by rheumatoid arthritis-associated transcriptional factor RUNX1 and inflammatory cytokines. Drug Metab Dispos. 2007; 35:394–401. PMID: 17142562.

20. Jang GH, Kim TH, Choe Y, Ham A, Choi JH. Functional characterization of genetic variations in the MDR3 promoter. Biochem Biophys Res Commun. 2013; 430:1312–1318. PMID: 23261441.

21. Yoo YK, Ke X, Hong S, Jang HY, Park K, Kim S, Ahn T, Lee YD, Song O, Rho NY, Lee MS, Lee YS, Kim J, Kim YJ, Yang JM, Song K, Kimm K, Weir B, Cardon LR, Lee JE, Hwang JJ. Fine-scale map of encyclopedia of DNA elements regions in the Korean population. Genetics. 2006; 174:491–497. PMID: 16702437.

22. Inoue T, Kamiyama J, Sakai T. Sp1 and NF-Y synergistically mediate the effect of vitamin D(3) in the p27(Kip1) gene promoter that lacks vitamin D response elements. J Biol Chem. 1999; 274:32309–32317. PMID: 10542271.

23. Lützner N, De-Castro Arce J, Rösl F. Gene expression of the tumour suppressor LKB1 is mediated by Sp1, NF-Y and FOXO transcription factors. PLoS One. 2012; 7:e32590. PMID: 22412893.

24. Bungartz G, Land H, Scadden DT, Emerson SG. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood. 2012; 119:1380–1389. PMID: 22072554.

25. Bernadt CT, Nowling T, Wiebe MS, Rizzino A. NF-Y behaves as a bifunctional transcription factor that can stimulate or repress the FGF-4 promoter in an enhancer-dependent manner. Gene Expr. 2005; 12:193–212. PMID: 16128003.
26. Ceribelli M, Dolfini D, Merico D, Gatta R, Viganò AM, Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol. 2008; 28:2047–2058. PMID: 18212061.

27. Newman B, Wintle RF, van Oene M, Yazdanpanah M, Owen J, Johnson B, Gu X, Amos CI, Keystone E, Rubin LA, Siminovitch KA. SLC22A4 polymorphisms implicated in rheumatoid arthritis and Crohn's disease are not associated with rheumatoid arthritis in a Canadian Caucasian population. Arthritis Rheum. 2005; 52:425–429. PMID: 15693005.

28. Barton A, Eyre S, Bowes J, Ho P, John S, Worthington J. Investigation of the SLC22A4 gene (associated with rheumatoid arthritis in a Japanese population) in a United Kingdom population of rheumatoid arthritis patients. Arthritis Rheum. 2005; 52:752–758. PMID: 15751072.
Fig. 1
Effect of variants on OCTN1 promoter activity. Promoter activities were measured 30 h after the transfection of reporter plasmids containing the major OCTN1 haplotypes (a) or genetic variants (b) into HCT-116 cells. The reporter activity of each construct was compared with empty vector (a, EV, pGL4.11b[luc2]) or the reference haplotype (b, H2). The data represent mean±SD of triplicate wells in a representative experiment. ***p<0.001.
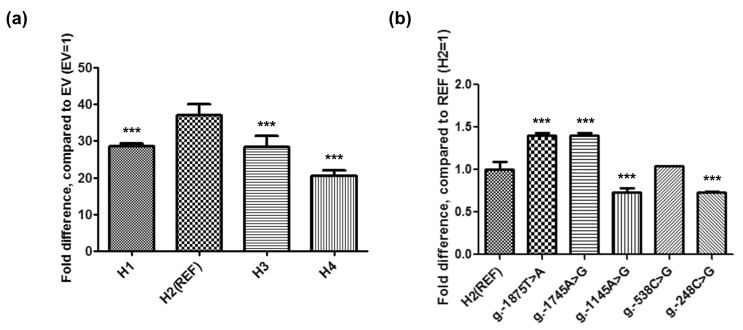
Fig. 2
Electrophoretic mobility shift analysis of the OCTN1 reference and g.-1875T>A variant sequences. (a) Labeled oligonucleotides (NF-Y consensus, lanes 1~3; OCTN1 reference, lanes 4~6; variant, lanes 7~9) were incubated with 35µg of nuclear protein extracts. Competition assays were performed using unlabeled NF-Y consensus (lanes 2, 5, and 8) or mutant (lanes 3, 6, and 9) oligonucleotides. The arrow indicates the position of the DNA-protein complex. (b) Labeled oligonucleotides (NF-Y consensus, lanes 1~3; OCTN1 reference, lane 4; variant, lane 5) were incubated with 20µg of nuclear protein extracts. Supershift assays were performed using a mixture of three different antibodies against NF-Y (lanes 2, 4, and 5) or an antibody against MZF-1 (lane 3).
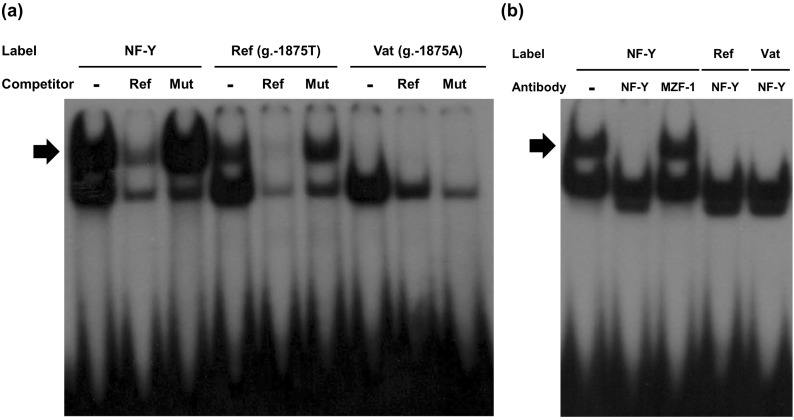
Fig. 3
Effect of NF-Y on OCTN1 promoter activity. Promoter activities were measured 30 h after the co-transfection of reference or variant reporters and varying amounts of NF-Y plasmids into HCT-116 cells. The reporter activity of each construct was compared with naïve promoter activity. The data represent mean±SD of triplicate wells in a representative experiment. *p<0.05, ***p<0.001 vs. naïve promoter activity and †p<0.01 vs. reference promoter activity.
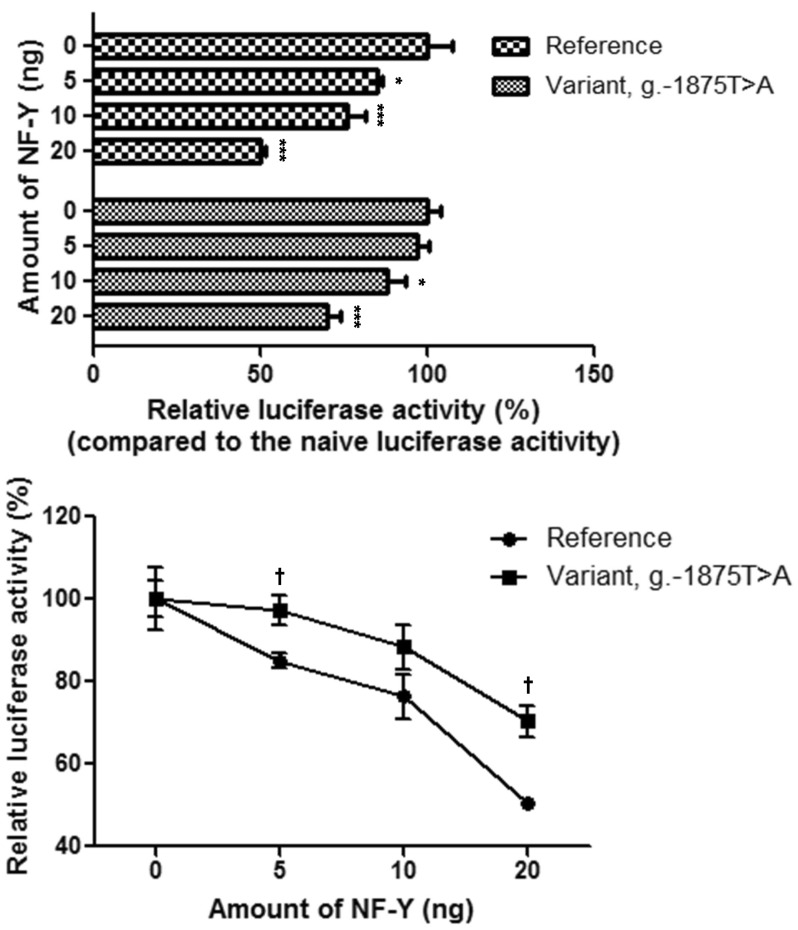
Table 1
Oligonucleotide primers used in the construction of OCTN1 reporter plasmids and electrophoretic mobility shift assays (EMSAs)

1The restriction endonuclease sites were marked by bold-faced letters with underlines. 2The SNP sites were marked by bold-faced letters with underlines. 3The consensus sequence of NF-Y was marked by bold-faced letters with underlines [21]. 4The changes in consensus sequences were marked by bold-faced letters with underlines.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download