INTRODUCTION
METHODS
Cell culture
Reagents
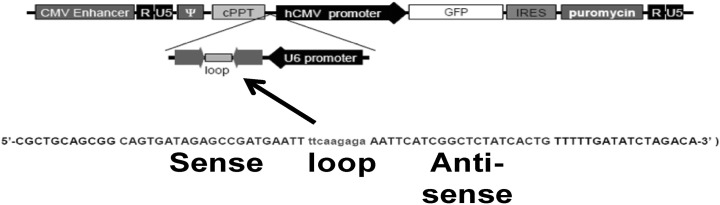
Establishment of the PRX-1 knockdown cell line
Cell viability assay
NO assay
Total RNA isolation, reverse transcription and polymerase chain reaction (RT-PCR)
Enzyme-linked immunosorbent assay (ELISA)
Fluorescence microscopy
Flow cytometry
Western blot analysis
Statistical analysis
RESULTS
Knockdown of PRX-1 through PRX-1 shLenti constructs in MIN6 and RAW264.7 cells
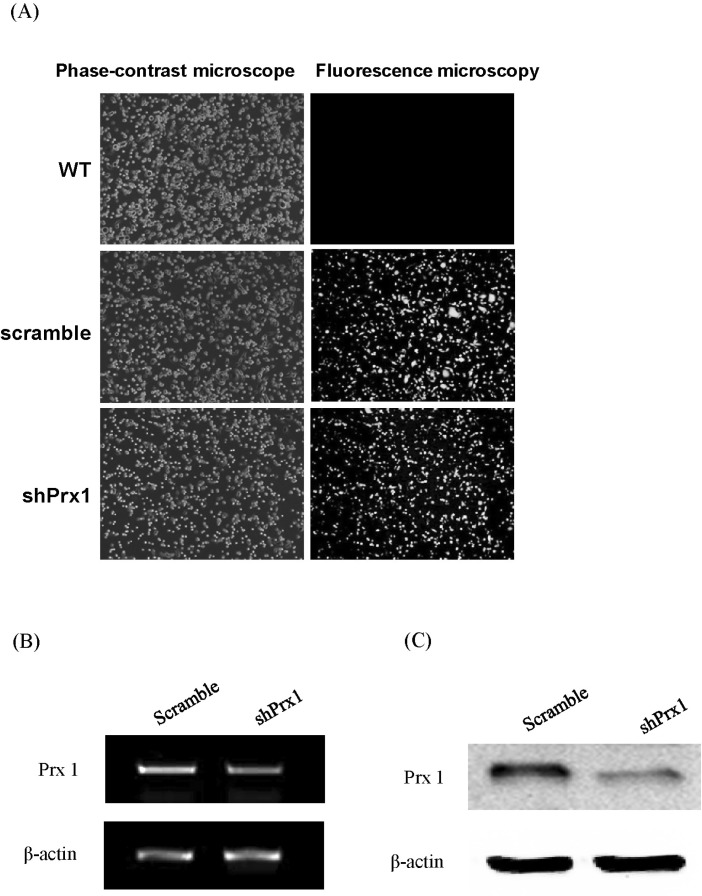 | Fig. 2(A) shPRX-1 construct and empty vector transduction efficiency. WT, scramble, and shPRX-1 1×106 MIN6 or RAW264.7 cells were harvested in PBS 1 ml. Then, the transduction efficiency was detected by phase fluorescence microscopy, representative imagery are shown. WT: wild type; Scramble: empty vector; shPRX-1: PRX-1-knockdown. (B) shPRX-1 silenced PRX-1 at gene level. Scramble, and shPRX-1 2×105 MIN6 or RAW264.7 cells were incubated in 24 well plate overnight. Total mRNA were extracted by Trizol and verified by RT-PCR, representative bands are shown. (C) shPRX-1 silenced PRX-1 at protein levels. Scramble, and shPRX-1 2×106 MIN6 or RAW264.7 cells were incubated in 6 well plate overnight. Whole protein was extracted by RIPA buffer and verified by western blot analysis, representative blots are shown. |
Determination of optimal condition of RAW264.7 cells for co-culture
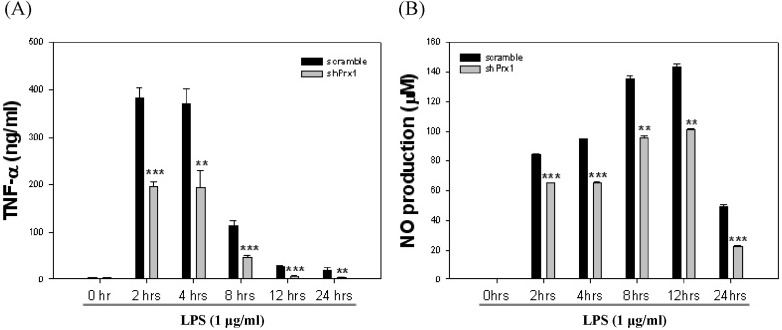 | Fig. 3Determination of optimal stimulation of RAW264.7 cells for co-culture. (A) RAW264.7 cells were stimulated with LPS (1 µg/ml) at varying time intervals from 0 hr to 24 hr, TNF-α levels were evaluated by ELISA at each time interval. Values represent the mean±SD (**, p<0.01 vs. scramble, ***, p<0.005 vs. scramble). (B) RAW264.7 cells were stimulated with LPS (1 µg/ml) at varying time intervals from 0 hr to 24 hr, nitric oxide (NO) levels were determined by NO assay at each time interval. Values represent the mean±SD (**, p<0.01 vs. scramble, ***, p<0.005 vs. scramble). |
MIN6 cells co-cultured with LPS-stimulated shPRX-1 RAW264.7 cells in the presence of L-NMMA showed increased anti-apoptotic gene expression compared to MIN6 cells co-cultured with RAW264.7 cells
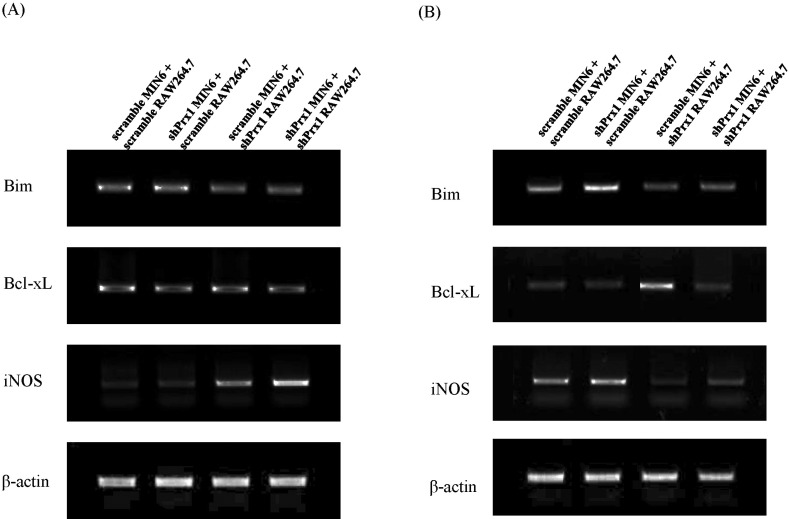 | Fig. 4Apoptotic gene expression in RAW264.7 and MIN6 co-cultures visualized through RT-PCR. (A) RAW264.7 cells were pre-stimulated with LPS for 4 hr before 24 hr of co-culture. Total RNA from each group was extracted and reverse transcribed for RT-PCR analysis. (B) Under the same conditions, L-NMMA was added to exclude the effect of NO and gene expression was assessed by RT-PCR. |
Decreased NO production is observed in MIN6 cells when treated with IL-6 and PRX-1 is required in the cell survival signals induced by IL-6
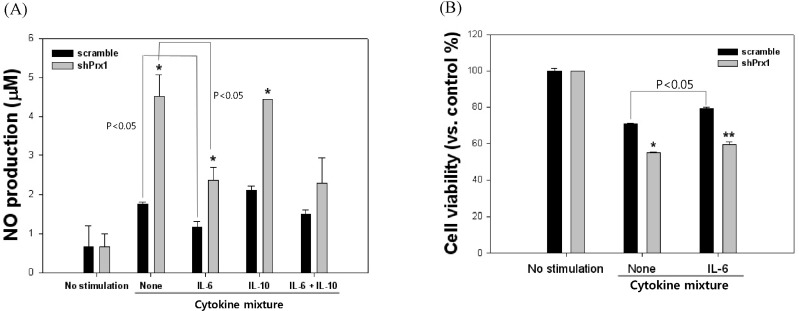 | Fig. 5NO production and cell viability of scramble and shPRX-1 MIN6 cells in the presence or absence of IL-6 and IL-10. (A) NO production of MIN6 cells were observed after treatment with an inflammatory cytokine mixture for 24 hr and the addition of IL-6, IL-10 or a combination of both. Values represent the mean±SD (*, p<0.05 vs. scramble, **, p<0.01 vs. scramble). (B) Cell viability of MIN6 cells were analyzed after treatment with an inflammatory cytokine mixture for 24 hr and in the presence or absence of IL-6. Values represent the mean±SD (*, p<0.05 vs. scramble, **, p<0.01 vs. scramble). |
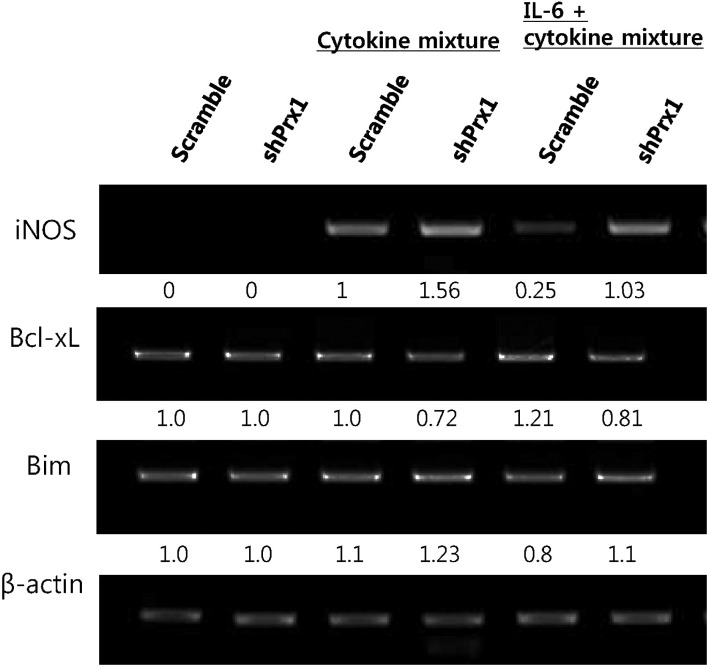 | Fig. 6The expression of iNOS, Bcl-xL and Bim was assessed by RT-PCR. Under cytokine mix treatment, IL-6 down-regulates iNOS expression in a significant manner in scramble MIN6 cells compared to shPRX-1 cells. shPRX-1 MIN6 cells treated with IL-6 showed increased expression of the anti-apoptotic gene, Bcl-xL and decreased expression of the pro-apoptotic gene, Bax. |
PRX-1 affects survival signals induced by IL-6 and also affects insulin production in MIN6 cells
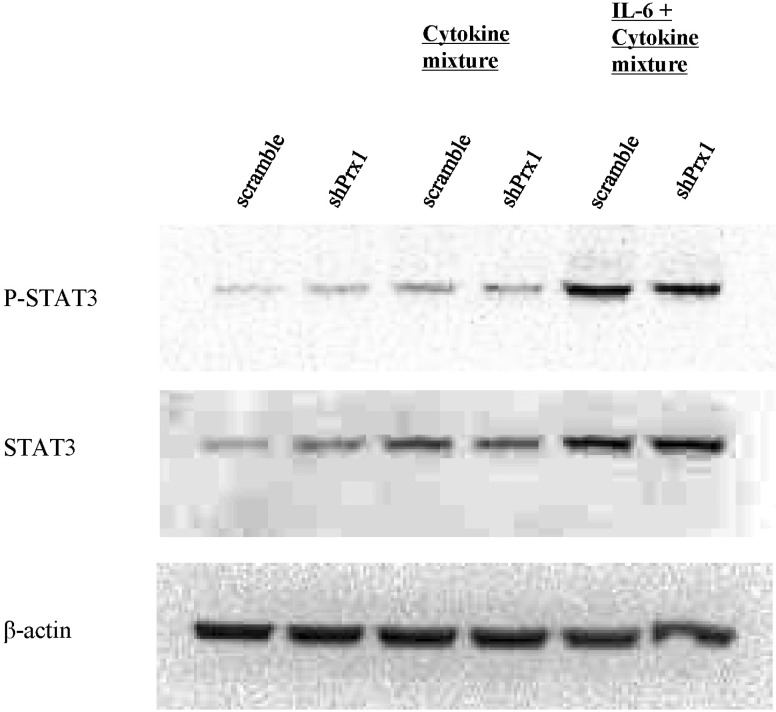 | Fig. 7PRX-1 promotes phosphorylation of STAT3. Whole protein was extracted through RIPA buffer and after performing SDS-PAGE electrophoresis, the protein was transferred to a PVDF membrane. The membrane was subsequently blocked and blots were visualized using antibodies for β-actin, phosphor-STAT3 and STAT3 and anti-mouse IgG or anti-rabbit IgG HRP-linked antibodies. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download