Abstract
Melatonin, which is the main product of the pineal gland, has well documented antioxidant and immune-modulatory effects. Macrophages produce molecules that are known to play roles in inflammatory responses. We conducted microarray analysis to evaluate the global gene expression profiles in response to treatment with melatonin in lipopolysaccharide (LPS) activated RAW 264.7 macrophage cells. In addition, eight genes were subjected to real-time reverse transcription polymerase chain reaction (RT-PCR) to confirm the results of the microarray. The cells were treated with LPS or melatonin plus LPS for 24 hr. LPS induced the up-regulation of 1073 genes and the down-regulation of 1144 genes when compared to the control group. Melatonin pretreatment of LPS-stimulated RAW 264.7 cells resulted in the down regulation of 241 genes and up regulation of 164 genes. Interestingly, among genes related to macrophage-mediated immunity, LPS increased the expression of seven genes (Adora2b, Fcgr2b, Cish, Cxcl10, Clec4n, Il1a, and Il1b) and decreased the expression of one gene (Clec4a3). These changes in expression were attenuated by melatonin. Furthermore, the results of real-time PCR were similar to those of the microarray. Taken together, these results suggest that melatonin may have a suppressive effect on LPS-induced expression of genes involved in the regulation of immunity and defense in RAW 264.7 macrophage cells. Moreover, these results may explain beneficial effects of melatonin in the treatment of various inflammatory conditions.
References
2. Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. 2009; 130:7–15.

3. Balkwill F, Coussens LM. Cancer: an inflammatory link. Nature. 2004; 431:405–406.
4. Consilvio C, Vincent AM, Feldman EL. Neuroinflammation, COX-2, and ALS–a dual role? Exp Neurol. 2004; 187:1–10.

6. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008; 8:958–969.

7. Kopydlowski KM, Salkowski CA, Cody MJ, van Rooijen N, Major J, Hamilton TA, Vogel SN. Regulation of macrophage chemokine expression by lipopolysaccharide in vitro and in vivo. J Immunol. 1999; 163:1537–1544.
8. Reiter RJ. Melatonin: clinical relevance. Best Pract Res Clin Endocrinol Metab. 2003; 17:273–285.

9. Tan DX, Reiter RJ, Manchester LC, Yan MT, El-Sawi M, Sainz RM, Mayo JC, Kohen R, Allegra M, Hardeland R. Chemical and physical properties and potential mechanisms: melatonin as a broad spectrum antioxidant and free radical scavenger. Curr Top Med Chem. 2002; 2:181–197.

10. Carrillo-Vico A, Lardone PJ, Naji L, Fernández-Santos JM, Martín-Lacave I, Guerrero JM, Calvo JR. Beneficial pleiotropic actions of melatonin in an experimental model of septic shock in mice: regulation of pro-/anti-inflammatory cytokine network, protection against oxidative damage and anti-apoptotic effects. J Pineal Res. 2005; 39:400–408.

11. Gitto E, Karbownik M, Reiter RJ, Tan DX, Cuzzocrea S, Chiurazzi P, Cordaro S, Corona G, Trimarchi G, Barberi I. Effects of melatonin treatment in septic newborns. Pediatr Res. 2001; 50:756–760.

12. Park HJ, Kim HJ, Ra J, Hong SJ, Baik HH, Park HK, Yim SV, Nah SS, Cho JJ, Chung JH. Melatonin inhibits lipopolysaccharide-induced CC chemokine subfamily gene expression in human peripheral blood mononuclear cells in a microarray analysis. J Pineal Res. 2007; 43:121–129.

13. Blackburn MR, Lee CG, Young HW, Zhu Z, Chunn JL, Kang MJ, Banerjee SK, Elias JA. Adenosine mediates IL-13-induced inflammation and remodeling in the lung and interacts in an IL-13-adenosine amplification pathway. J Clin Invest. 2003; 112:332–344.

14. Fredholm BB, Abbracchio MP, Burnstock G, Daly JW, Harden TK, Jacobson KA, Leff P, Williams M. Nomenclature and classification of purinoceptors. Pharmacol Rev. 1994; 46:143–156.
15. Reutershan J, Cagnina RE, Chang D, Linden J, Ley K. Therapeutic anti-inflammatory effects of myeloid cell adenosine receptor A2a stimulation in lipopolysaccharide-induced lung injury. J Immunol. 2007; 179:1254–1263.

16. Eckle T, Grenz A, Laucher S, Eltzschig HK. A2B adenosine receptor signaling attenuates acute lung injury by enhancing alveolar fluid clearance in mice. J Clin Invest. 2008; 118:3301–3315.

17. Schingnitz U, Hartmann K, Macmanus CF, Eckle T, Zug S, Colgan SP, Eltzschig HK. Signaling through the A2B adenosine receptor dampens endotoxin-induced acute lung injury. J Immunol. 2010; 184:5271–5279.

18. Takai T. Fc receptors and their role in immune regulation and autoimmunity. J Clin Immunol. 2005; 25:1–18.

19. Jin HJ, Shao JZ, Xiang LX, Wang H, Sun LL. Global identification and comparative analysis of SOCS genes in fish: insights into the molecular evolution of SOCS family. Mol Immunol. 2008; 45:1258–1268.

20. Kim BH, Lee KH, Chung EY, Chang YS, Lee H, Lee CK, Min KR, Kim Y. Inhibitory effect of chroman carboxamide on interleukin-6 expression in response to lipopolysaccharide by preventing nuclear factor-kappaB activation in macrophages. Eur J Pharmacol. 2006; 543:158–165.
21. Feferman T, Maiti PK, Berrih-Aknin S, Bismuth J, Bidault J, Fuchs S, Souroujon MC. Overexpression of IFN-induced protein 10 and its receptor CXCR3 in myasthenia gravis. J Immunol. 2005; 174:5324–5331.

22. Lord PC, Wilmoth LM, Mizel SB, McCall CE. Expression of interleukin-1 alpha and beta genes by human blood polymorphonuclear leukocytes. J Clin Invest. 1991; 87:1312–1321.

23. Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, Wilson TG Jr, Higginbottom FL, Duff GW. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997; 24:72–77.

24. Asensi V, Alvarez V, Valle E, Meana A, Fierer J, Coto E, Carton JA, Maradona JA, Paz J, Dieguez MA, de la Fuente B, Moreno A, Rubio S, Tuya MJ, Sarasúa J, Llames S, Arribas JM. IL-1 alpha (-889) promoter polymorphism is a risk factor for osteomyelitis. Am J Med Genet A. 2003; 119A:132–136.
25. Mwantembe O, Gaillard MC, Barkhuizen M, Pillay V, Berry SD, Dewar JB, Song E. Ethnic differences in allelic associations of the interleukin-1 gene cluster in South African patients with inflammatory bowel disease (IBD) and in control individuals. Immunogenetics. 2001; 52:249–254.
Fig. 1.
Cytotoxicity of melatonin was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. RAW 264.7 macrophage cells were exposed to melatonin at 0, 50, 100, and 500 μM in the absence or presence of LPS for 24 h. The results are presented as the mean±S.E.M. LPS, lipopolysaccharide.
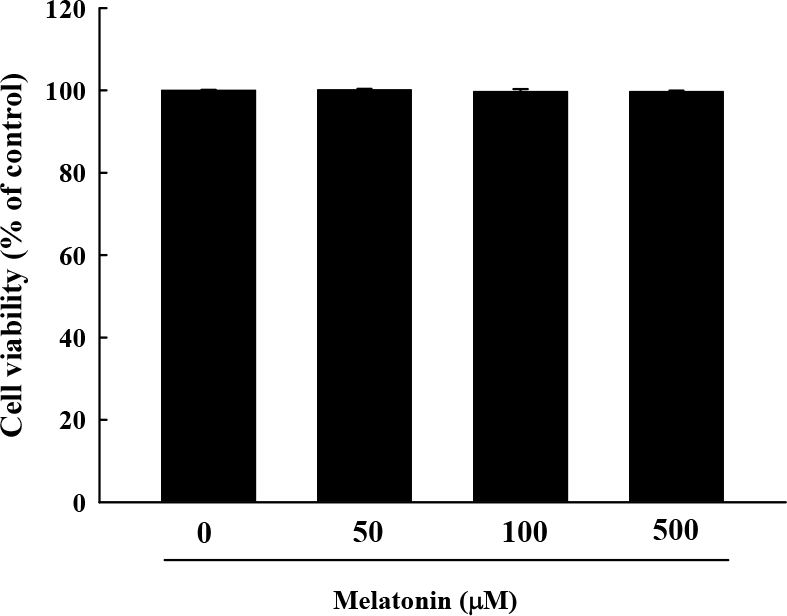
Fig. 2.
The effects of melatonin on nitrite production. RAW 264.7 cells were pretreated with 50, 100, and 500 μM of melatonin and then treated with lipopolysaccharides (1.0 μg/ml). The media were harvested 24 h later and assayed for nitrite production. Data are the means±S.E.M. LPS, lipopolysaccharide; Mel, melatonin. ##p< 0.01 compared to control, ∗∗p<0.01 compared to LPS.
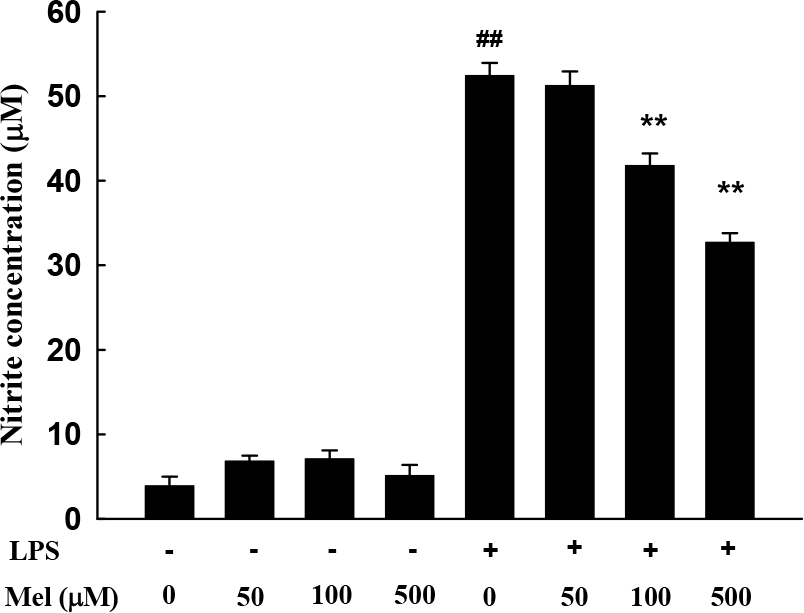
Fig. 3.
Functional analysis of genes selected using the Panther database. (A) Functional categories of genes regulated by LPS treatment in RAW 264.7 macrophage cells. (B) Functional categories of genes regulated by melatonin pretreatment in LPS-stimulated RAW 264.7 macrophage cells.
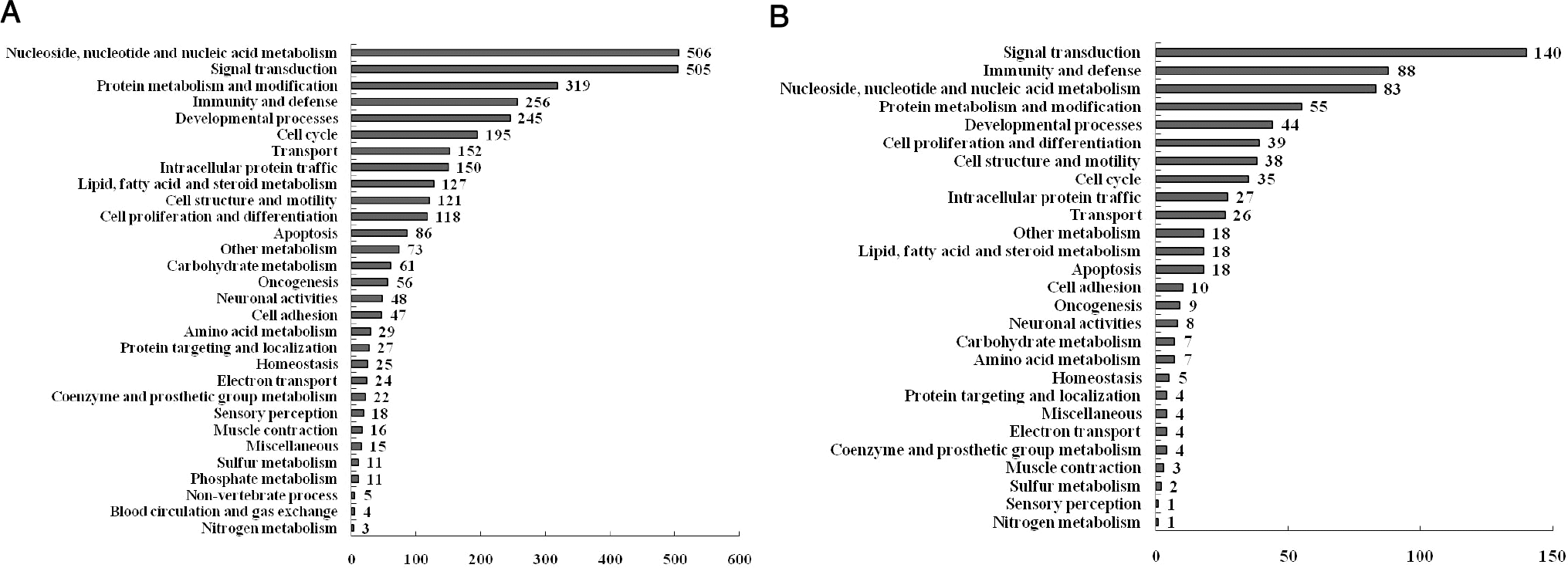
Fig. 4.
Validation of microarray results using real-time RT-PCR. Relative expression of each gene in the control was designated as 1, values are mean±S.E.M. LPS, lipopolysaccharide; Mel, melatonin. #p<0.05, ##p<0.01 compared to control, ∗p<0.05, ∗∗p<0.01 compared to LPS.
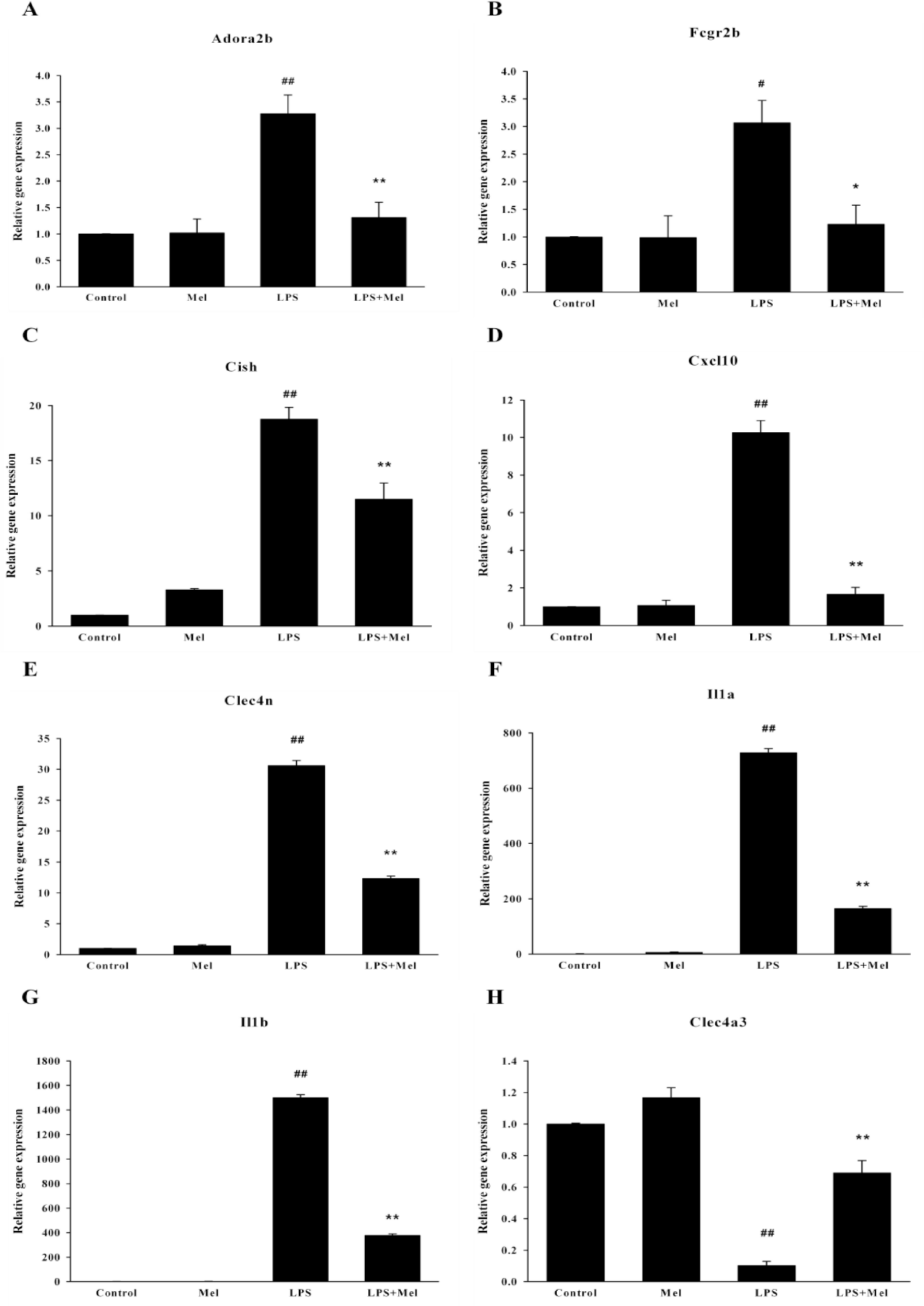
Table 1.
Primer sequences of genes applied in real-time RT-PCR
Table 2.
List of “macrophage-mediated immunity” genes regulated by lipopolysaccharide treatment in RAW 264.7 macrophage cells
Table 3.
List of “macrophage-mediated immunity” genes regulated by melatonin pretreatment in lipopolysaccharide-stimulated RAW 264.7 macrophage cells




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download