Abstract
Amyloid precursor protein binding protein-1 (APP-BP1) binds to the carboxyl terminus of amyloid precursor protein and serves as a bipartite activation enzyme for the ubiquitin-like protein, NEDD8. Previously, it has been reported that APP-BP1 rescues the cell cycle S-M checkpoint defect in Ts41 hamster cells, that this rescue is dependent on the interaction of APP-BP1 with hUba3. The exogenous expression of APP-BP1 in neurons has been reported to cause DNA synthesis and apoptosis via a signaling pathway that is dependent on APP-BP1 binding to APP. These results suggest that APP-BP1 overexpression contributes to neurodegeneration. In the present study, we explored whether APP-BP1 expression was altered in the brains of Tg2576 mice, which is an animal model of Alzheimer's disease. APP-BP1 was found to be up-regulated in the hippocampus and cortex of 12 month-old Tg2576 mice compared to age-matched wild-type mice. In addition, APP-BP1 knockdown by siRNA treatment reduced cullin-1 neddylation in fetal neural stem cells, suggesting that APP-BP1 plays a role in cell cycle progression in the cells. Collectively, these results suggest that increased expression of APP-BP1, which has a role in cell cycle progression in neuronal cells, contributes to the pathogenesis of Alzheimer's disease.
Go to : 
References
1. Chen Y, Liu W, McPhie DL, Hassinger L, Neve RL. APP-BP1 mediates APP-induced apoptosis and DNA synthesis and is increased in Alzheimer's disease brain. J Cell Biol. 2003; 163:27–33.

2. Chow N, Korenberg JR, Chen XN, Neve RL. APP-BP1, a novel protein that binds to the carboxyl-terminal region of the amyloid precursor protein. J Biol Chem. 1996; 271:11339–11346.

3. Selkoe DJ. Soluble oligomers of the amyloid beta-protein impair synaptic plasticity and behavior. Behav Brain Res. 2008; 192:106–113.
4. Bruni P, Minopoli G, Brancaccio T, Napolitano M, Faraonio R, Zambrano N, Hansen U, Russo T. Fe65, a ligand of the Alzheimer's beta-amyloid precursor protein, blocks cell cycle progression by down-regulating thymidylate synthase expression. J Biol Chem. 2002; 277:35481–35488.
5. Tang BL, Liou YC. Novel modulators of amyloid-β precursor protein processing. J Neurochem. 2007; 100:314–323.
6. Sakuma M, Tanaka E, Taru H, Tomita S, Gandy S, Nairn AC, Nakaya T, Yamamoto T, Suzuki T. Phosphorylation of the amino-terminal region of X11L regulates its interaction with APP. J Neurochem. 2009; 109:465–475.

7. Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999. 274,12036.

8. Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999; 18:6829–6834.

9. Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998; 12:2263–2268.
12. Kamitani TK, Kito HP, Nguyen , Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997; 272:28557–28562.

13. Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001; 155:571–579.

14. Kitahara R, Yamaguchi Y, Sakata E, Kasuya T, Tanaka K, Kato K, Yokoyama S, Akasaka K. Evolutionally conserved intermediates between ubiquitin and NEDD8. J Mol Biol. 2006; 363:395.

15. Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009; 66:1924–1938.

16. Ohki Y, Funatsu N, Konishi N, Chiba T. The mechanism of poly-NEDD8 chain formation in vitro. Biochem Biophys Res Commun. 2009; 381:443–447.

17. Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. Cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996; 85:829–839.

19. Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001; 3:321–324.

20. Bassermann F, Pagano M. Dissecting the role of ubiquitylation in the DNA damage response checkpoint in G2. Cell Death Differ. 2010; 17:78–85.

21. Cardozo T, Pagano M. Wrenches in the works: drug discovery targeting the SCF ubiquitin ligase and APC/C complexes. BMC Biochem. 2007. 8 Suppl 1:S9.

22. Chen Y, McPhie DL, Hirschberg J, Neve RL. The Amyloid Precursor Protein-binding Protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J Biol Chem. 2000; 275:8929–8935.

23. Chen Y, Liu W, Naumovski L, Neve RL. ASPP2 inhibits APP-BP1-mediated NEDD8 conjugation to cullin-1 and decreases APP-BP1-induced cell proliferation and neuronal apoptosis. J Neurochem. 2003; 85:801–809.

24. Laifenfeld D, Patzek LJ, McPhie DL, Chen Y, Levites Y, Cataldo AM, Neve RL. Rab5 Mediates an Amyloid Precursor Protein Signaling Pathway That Leads to Apoptosis. J Neurosci. 2007; 27:7141–7153.

26. Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996; 10:3129–3140.

27. Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992; 12:4565–4574.

28. Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science. 1996; 274:99–102.
29. Laemili UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227:680–685.
30. Zekanowski C, Wojda U. Aneuploidy, chromosomal missegragation, and cell cycle reentry in Alzheimeri C and Wojda U. Aneuploidy, chromosomal missegregation, and cell cycle reentry in Alzheimer's disease. Acta Neurobiol Exp (Wars). 2009; 69:232–253.
31. Bonda DJ, Lee HP, Kudo W, Zhu X, Smith MA, Lee HG. Pathological implications of cell cycle re-entry in Alzheimer's disease. Expert Rev Mol Med. 2010; 12:e19.
32. Rothman SM, Mattson MP. Adverse stress, hippocampal networks, and Alzheimer's disease. Neuromolecular Med. 2010; 12:56–70.

33. Park Y, Yoon SK, Yoon JB. TRIP12 functions as an E3 ubiquitin ligase of APP-BP1. Biochem Biophys Res Commun. 2008; 374:294–298.

34. Walden H, Podgorski , Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003; 422:330–334.

35. Gustaw-Rothenberg K, Lerner A, Bonda DJ, Lee HG, Zhu X, Perry G, Smith MA. Biomarkers in Alzheimer's disease: past, present and future. Biomark Med. 2010; 4:15–26.

36. Yang Y, Herrup K. Cell division in the CNS: Protective response or lethal event in post-mitotic neurons? Biochim Biophy Acta. 2007; 1772:457–466.

37. Mcshera A, Wahl AF, Smith MA. RE-entry into the cell cycle: a mechanism for neurodegeneration in Alzheimer's disease. Medical Hypotheses. 1999; 52:525–527.
38. Nagy Z, Esiri MM, Cato AM, Smith AD. Cell cycle markers in the hippocampus in Alzheimer's disease. Acta Neuropathol. 1997; 94:6–15.

39. Neve RL, McPhie DL. Dysfunction of amyloid precursor protein signaling in neurons leads to DNA synthesis and apoptosis. Biochim Biophys Acta. 2007; 1772:430–437.

40. Bowser R, Smith MA. Cell cycle proteins in Alzheimer's disease: plenty of wheels but no cycle. J Alzheimers Dis. 2002; 4:249–254.

41. Herrup R, Neve SL, Ackerman A, Copani A. Divide and die: cell cycle events as triggers of nerve cell death. J Neurosci. 2004; 24:9232–9239.

42. Zhu X, McShea A, Harris PLR, Raina AK, Castellani RJ, Funk JO, Shah S, Atwood C, Bowen R, Bowser R, Morelli L, Perry G, Smith MA. Elevated expression of a regulator of the G2/M phase of the cell cycle, neuronal CIP-1-associated regulator of cyclin B, in Alzheimer's disease. J Neurosci Res. 2004; 75:698–703.

Go to : 
 | Fig. 1.APP-BP1 protein level was up-regulated in the hippocampus from 12 month-old Tg2576 mice. (A) APP-BP1 protein level was examined by western blotting in the hippocampus from 6-, 12- and 18-month-old Tg 2576 and wild-type mice. This blot is a representative of 4 independent experiments. (B) Densitometric analysis was also performed. Data represent mean±SEM. ∗p<0.05 compared with 6 month-old wild type mice (n=4) and #p<0.05 compared with 12 month-old wild type mice, by one-way ANOVA followed by Duncan's post hoc test. |
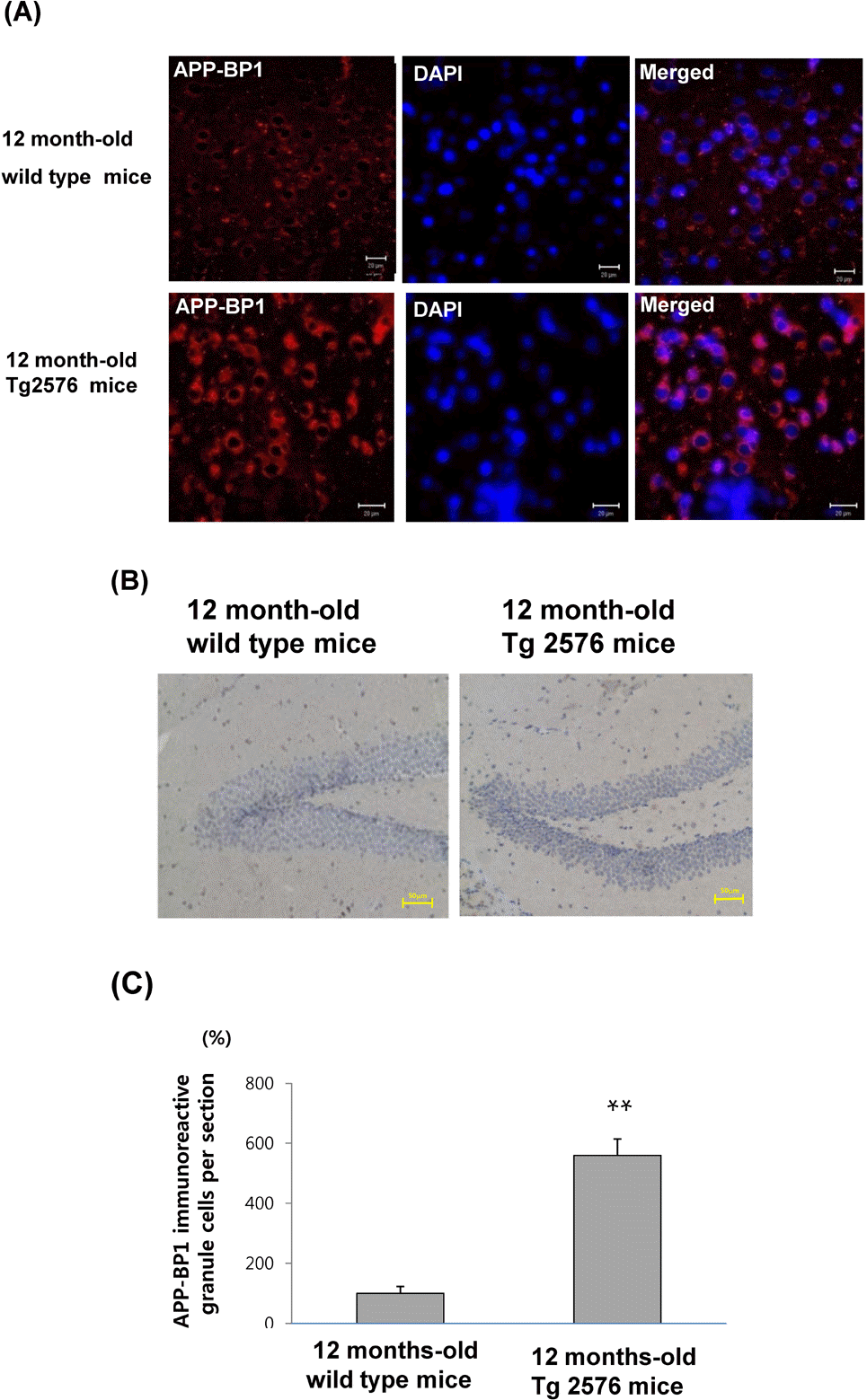 | Fig. 2.APP-BP1 immunoreactivities are increased in the cortex and hippocampus from 12-month-old wild-type and Tg2576 mice. The fixed brains from 12-month-old wild type and Tg2576 mice (n=5) in 10% neutral buffered formalin for 48 h were dehydrated and embedded in paraffin. (A) The fluorescent immunohistochemistry was performed in the cortex with the anti-APP-BP1 antibody for 2 h, which was then visualized using a Cy3-conjugated secondary antibody. DAPI counter staining was performed. Images were collected using the LSM 510 program on a Zeiss confocal microscope. This figure is a representative of 5 independent experiments. Scale bars=25 μm. (B) Immunoreactivities of APP-BP1 was examined in the dentate gyrus of 12 month-old wild type and Tg2576 mice. IHC was performed using a Vectastain avidin-biotin complex elite kit. Peroxidase stained sections were examined under a light microscope (BX51TF; Olympus Optical Co., Japan) equipped with an exposure control unit (PM-CB20) and a camera (CPM-C35DX). Scale bars=50μm. (C) Quantative analysis for (B) was performed by calculating APP-BP1 immunoreactive cells per section from 5 independent samples (∗∗p<0.01 by Student's t-test). |
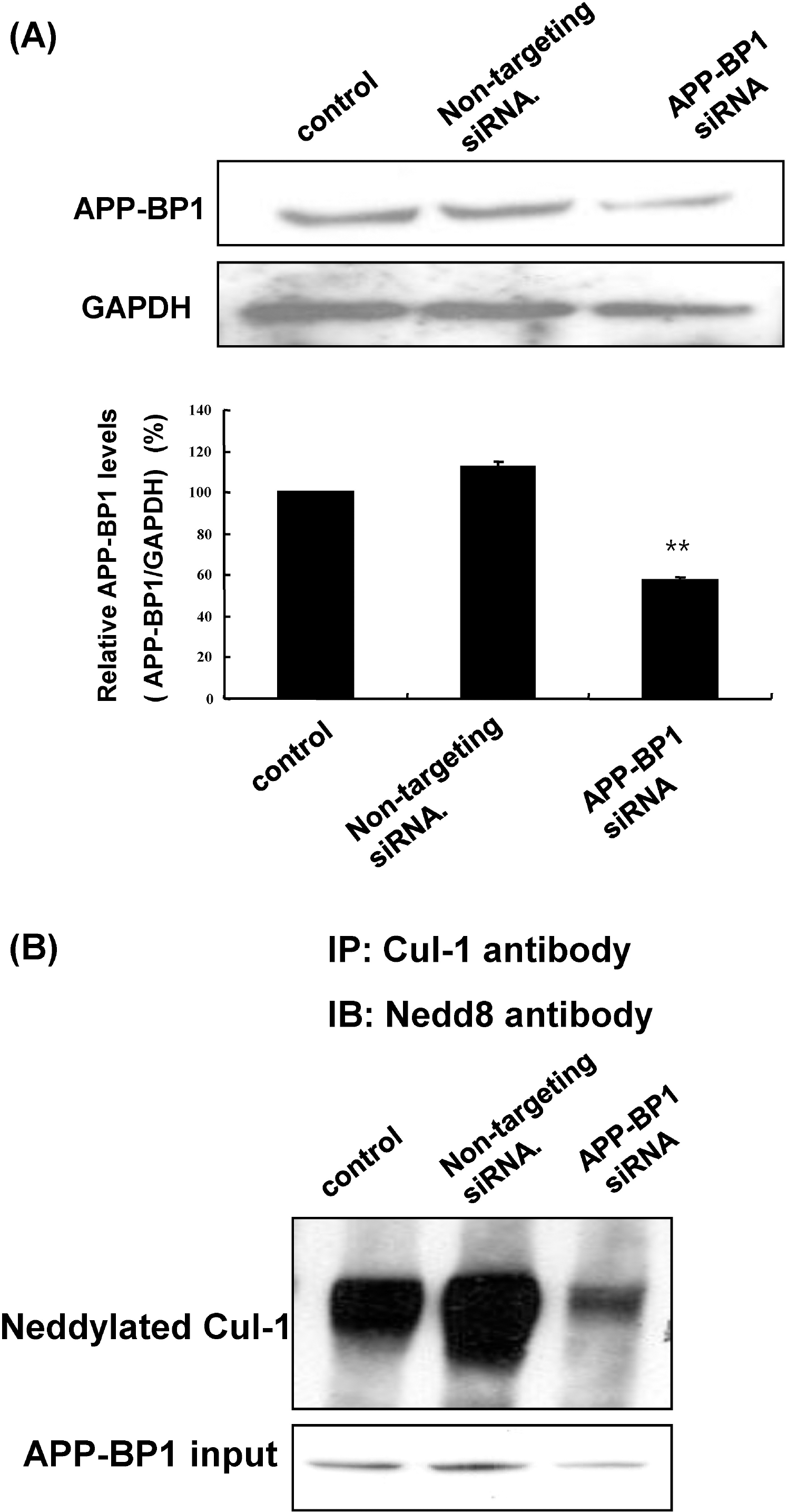 | Fig. 3.APP-BP1 knockdown reduced Cul-1 neddylation in fetal neural stem cells. (A) After treatment of fetal neural stem cells with 10 nM non-targeting and APP-BP1 siRNAs for 72 h, APP-BP1 protein levels were examined by western blotting. Densitometric analysis was also performed. Data represent mean±SEM. ∗p<0.05 compared with non-treated cells, by one-way ANOVA followed by Duncan's post hoc test. (B) After treatment with siRNAs in fetal neural stem cells (passage 8) for 72 h, neddylated Cul-1 was examined by immunoprecipitation of Cul-1 followed by western blotting for NEDD8. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download