Abstract
The tubby mouse is characterized by progressive retinal and cochlear degeneration and late-onset obesity. These phenotypes are caused by a loss-of-function mutation in the tub gene and are shared with several human syndromes, suggesting the importance of tubby protein in central nervous system (CNS) functioning. Although evidence suggests that tubby may act as a transcription factor mediating G-protein coupled receptor (GPCR) signaling, any downstream gene regulated by tubby has yet to be identified. To explore potential target genes of tubby with region-specific transcription patterns in the brain, we performed a microarray analysis using the cerebral cortex and hypothalamus of tubby mice. We also validated the changes of gene expression level observed with the microarray analysis using real-time RT-PCR. We found that expression of erythroid differentiation factor 1 (Erdr1) and caspase 1 (Casp1) increased, while p21-activated kinase 1 (Pak1) and cholecystokinin 2 receptor (Cck2r) expression decreased in the cerebral cortex of tubby mice. In the hypothalamic region, Casp 1 was up-regulated and μ-crystallin (CRYM) was down-regulated. Based on the reported functions of the differentially expressed genes, these individual or grouped genes may account for the phenotype of tubby mice. We discussed how altered expression of genes in tubby mice might be understood as the underlying mechanism behind tubby phenotypes.
Go to : 
REFERENCES
Abe S., Katagiri T., Saito-Hisaminato A., Usami S., Inoue Y., Tsunoda T., Nakamura Y. Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am J Hum Genet. 72:73–82. 2003.
Bode C., Wolfrum U. Caspase-3 inhibitor reduces apototic photoreceptor cell death during inherited retinal degeneration in tubby mice. Mol Vis. 9:144–150. 2003.
Boggon TJ., Shan WS., Santagata S., Myers SC., Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 286:2119–2125. 1999.

Cau J., Hall A. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J Cell Sci. 118:2579–2587. 2005.

Clerc P., Coll Constans MG., Lulka H., Broussaud S., Guigne C., Leung-Theung-Long S., Perrin C., Knauf C., Carpene C., Penicaud L., Seva C., Burcelin R., Valet P., Fourmy D., Dufresne M. Involvement of cholecystokinin 2 receptor in food intake regulation: hyperphagia and increased fat deposition in cholecystokinin 2 receptor-deficient mice. Endocrinology. 148:1039–1049. 2007.

Coleman DL., Eicher EM. Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered. 81:424–427. 1990.

Dormer P., Spitzer E., Moller W. EDR is a stress-related survival factor from stroma and other tissues acting on early haematopoietic progenitors (E-Mix). Cytokine. 27:47–57. 2004a.

Dormer P., Spitzer E., Frankenberger M., Kremmer E. Erythroid differentiation regulator (EDR), a novel, highly conserved factor I. Induction of haemoglobin synthesis in erythroleukaemic cells. Cytokine. 26:231–242. 2004b.
Gaudreau P., Quirion R., St-Pierre S., Pert CB. Characterization and visualization of cholecystokinin receptors in rat brain using [3H]pentagastrin. Peptides. 4:755–762. 1983.

Gillingwater TH., Wishart TM., Chen PE., Haley JE., Robertson K., MacDonald SH., Midleton S., Wawrowski K., Shipston MJ., Melmed S., Wyllie DJ., Skehel PA., Coleman MP., Ribchester RR. The neuroprotective WldS gene regulates expression of PTTG1 and erythroid differentiation regulator 1-like gene in mice and human cells. Hum Mol Genet. 15:625–635. 2006.

Guan XM., Yu H., Van der Ploeg LH. Evidence of altered hypothalamic pro-opiomelanocortin/neuropeptide Y mRNA expression in tubby mice. Brain Res Mol Brain Res. 59:273–279. 1998.
Hill DR., Shaw TM., Woodruff GN. Species differences in the localization of ‘peripheral’ type cholecystokinin receptors in rodent brain. Neurosci Lett. 79:286–289. 1987a.

Hill DR., Campbell NJ., Shaw TM., Woodruff GN. Autoradiographic localization and biochemical characterization of peripheral type CCK receptors in rat CNS using highly selective nonpeptide CCK antagonists. J Neurosci. 7:2967–2976. 1987b.

Hing H., Xiao J., Harden N., Lim L., Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 97:853–863. 1999.

Ikeda A., Naggert JK., Nishina PM. Genetic modification of retinal degeneration in tubby mice. Exp Eye Res. 74:455–461. 2002.

Ikeda S., He W., Ikeda A., Naggert JK., North MA., Nishina PM. Cell-specific expression of tubby gene family members (tub, Tulp1,2, and 3) in the retina. Invest Ophthalmol Vis Sci. 40:2706–2712. 1999.
Jacobs T., Causeret F., Nishimura YV., Terao M., Norman A., Hoshino M., Nikolic M. Localized activation of p21-activated kinase controls neuronal polarity and morphology. J Neurosci. 27:8604–8615. 2007.

Katsanis N., Lupski JR., Beales PL. Exploring the molecular basis of Bardet-Biedl syndrome. Hum Mol Genet. 10:2293–2299. 2001.

Kleyn PW., Fan W., Kovats SG., Lee JJ., Pulido JC., Wu Y., Berkemeier LR., Misumi DJ., Holmgren L., Charlat O., Woolf EA., Tayber O., Brody T., Shu P., Hawkins F., Kennedy B., Baldini L., Ebeling C., Alperin GD., Deeds J., Lakey ND., Culpepper J., Chen H., Glücksmann-Kuis MA., Carlson GA., Duyk GM., Moore KJ. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 85:281–290. 1996.

Koritschoner N., Alvarez-Dolado M., Kurz S., Heikenwälder M., Hacker C., Vogel F., Muñoz A., Zenke M. Thyroid hormone regulates the obesity gene tub. EMBO Rep. 2:499–504. 2001.
Kremer H., van Wijk E., Marker T., Wolfrum U., Roepman R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum Mol Genet 15 Spec No. 2:R262–270. 2006.

Lee JH., Lee J., Seo GH., Kim CH., Ahn YS. Heparin inhibits NF-kappaB activation and increases cell death in cerebral endothelial cells after oxygen-glucose deprivation. J Mol Neurosci. 32:145–154. 2007.
Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 10:417–426. 2002.
Morency MA., Quirion R., Mishra RK. Distribution of cholecy-stokinin receptors in the bovine brain: a quantitative autoradiographic study. Neuroscience. 62:307–316. 1994.

O'Shea E., Sanchez V., Orio L., Escobedo I., Green AR., Colado MI. 3,4-Methylenedioxymethamphetamine increases pro-interleukin-1beta production and caspase-1 protease activity in frontal cortex, but not in hypothalamus, of Dark Agouti rats: role of interleukin-1beta in neurotoxicity. Neuroscience. 135:1095–1105. 2005.
Ohlemiller KK., Hughes RM., Mosinger-Ogilvie J., Speck JD., Grosof DH., Silverman MS. Cochlear and retinal degeneration in the tubby mouse. Neuroreport. 6:845–849. 1995.

Orio L., O'Shea E., Sanchez V., Pradillo JM., Escobedo I., Camarero J., Moro MA., Green AR., Colado MI. 3,4-Methylenedioxy-methamphetamine increases interleukin-1beta levels and activates microglia in rat brain: studies on the relationship with acute hyperthermia and 5-HT depletion. J Neurochem. 89:1445–1453. 2004.
Oshima A., Suzuki S., Takumi Y., Hashizume K., Abe S., Usami S. CRYM mutations cause deafness through thyroid hormone binding properties in the fibrocytes of the cochlea. J Med Genet. 43:e25. 2006.

Perche O., Doly M., Ranchon-Cole I. Caspase-dependent apoptosis in light-induced retinal degeneration. Invest Ophthalmol Vis Sci. 48:2753–2759. 2007.

Roberts MR., Srinivas M., Forrest D., Morreale de Escobar G., Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci U S A. 103:6218–6223. 2006.

Santagata S., Boggon TJ., Baird CL., Gomez CA., Zhao J., Shan WS., Myszka DG., Shapiro L. G-protein signaling through tubby proteins. Science. 292:2041–2050. 2001.

Schurmann A., Mooney AF., Sanders LC., Sells MA., Wang HG., Reed JC., Bokoch GM. p21-activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol. 20:453–461. 2000.

Segovia L., Horwitz J., Gasser R., Wistow G. Two roles for mu-crystallin: a lens structural protein in diurnal marsupials and a possible enzyme in mammalian retinas. Mol Vis. 3:9. 1997.
Sells MA., Knaus UG., Bagrodia S., Ambrose DM., Bokoch GM., Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 7:202–210. 1997.

Stubdal H., Lynch CA., Moriarty A., Fang Q., Chickering T., Deeds JD., Fairchild-Huntress V., Charlat O., Dunmore JH., Kleyn P., Huszar D., Kapeller R. Targeted deletion of the tub mouse obesity gene reveals that tubby is a loss-of-function mutation. Mol Cell Biol. 20:878–882. 2000.
Suzuki S., Mori J., Hashizume K. Mu-crystallin, a NADPH-dependent T(3)-binding protein in cytosol. Trends Endocrinol Metab. 18:286–289. 2007.
Suzuki S., Mori J., Kobayashi M., Inagaki T., Inaba H., Komatsu A., Yamashita K., Takeda T., Miyamoto T., Ichikawa K., Hashizume K. Cell-specific expression of NADPH-dependent cytosolic 3,5,3'-triiodo-L-thyronine-binding protein (p38CTBP). Eur J Endocrinol. 148:259–268. 2003.

Tang Y., Zhou H., Chen A., Pittman RN., Field J. The Akt proto-oncogene links Ras to Pak and cell survival signals. J Biol Chem. 275:9106–9109. 2000.

Umeda S., Suzuki MT., Okamoto H., Ono F., Mizota A., Terao K., Yoshikawa Y., Tanaka Y., Iwata T. Molecular composition of drusen and possible involvement of anti-retinal autoimmunity in two different forms of macular degeneration in cynomolgus monkey (Macaca fascicularis). FASEB J. 19:1683–1685. 2005.
Wang Y., Barbacioru C., Hyland F., Xiao W., Hunkapiller KL., Blake J., Chan F., Gonzalez C., Zhang L., Samaha RR. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 7:59. 2006.

Zapala MA., Hovatta I., Ellison JA., Wodicka L., Del Rio JA., Tennant R., Tynan W., Broide RS., Helton R., Stoveken BS., Winrow C., Lockhart DJ., Reilly JF., Young WG., Bloom FE., Barlow C. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 102:10357–10362. 2005.

Go to : 
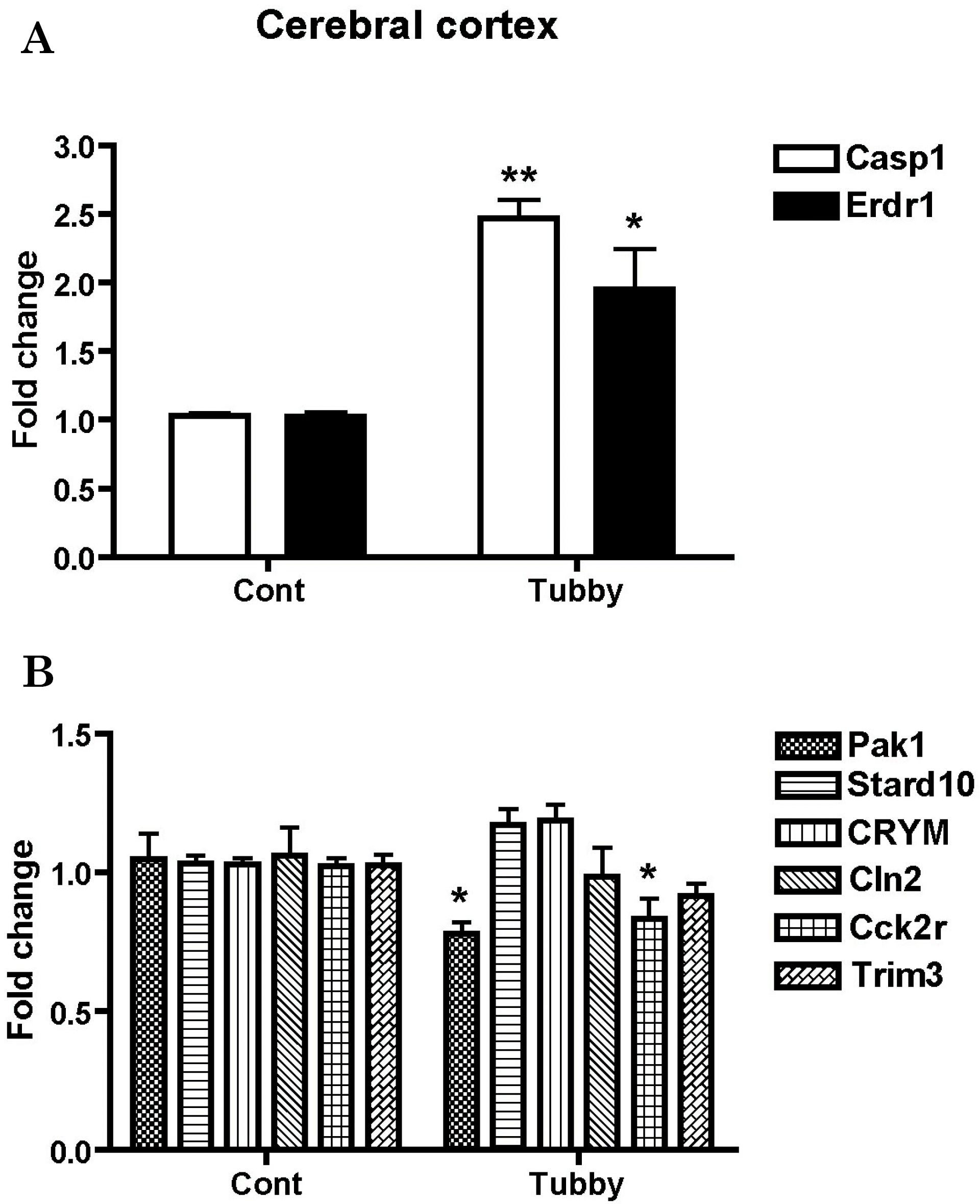 | Fig. 1.Validation of array-based gene expression profiles in the cerebral cortex of tubby mice using real-time RT-PCR. mRNA levels of genes were determined using real-time RT-PCR. Expression profiles of up-regulated (A) and down-regulated genes (B) are presented as fold changes compared to the cerebral cortex of control mice. Expression levels of β-actin were used to normalize the values. ∗p<0.05 and ∗∗p<0.01. |
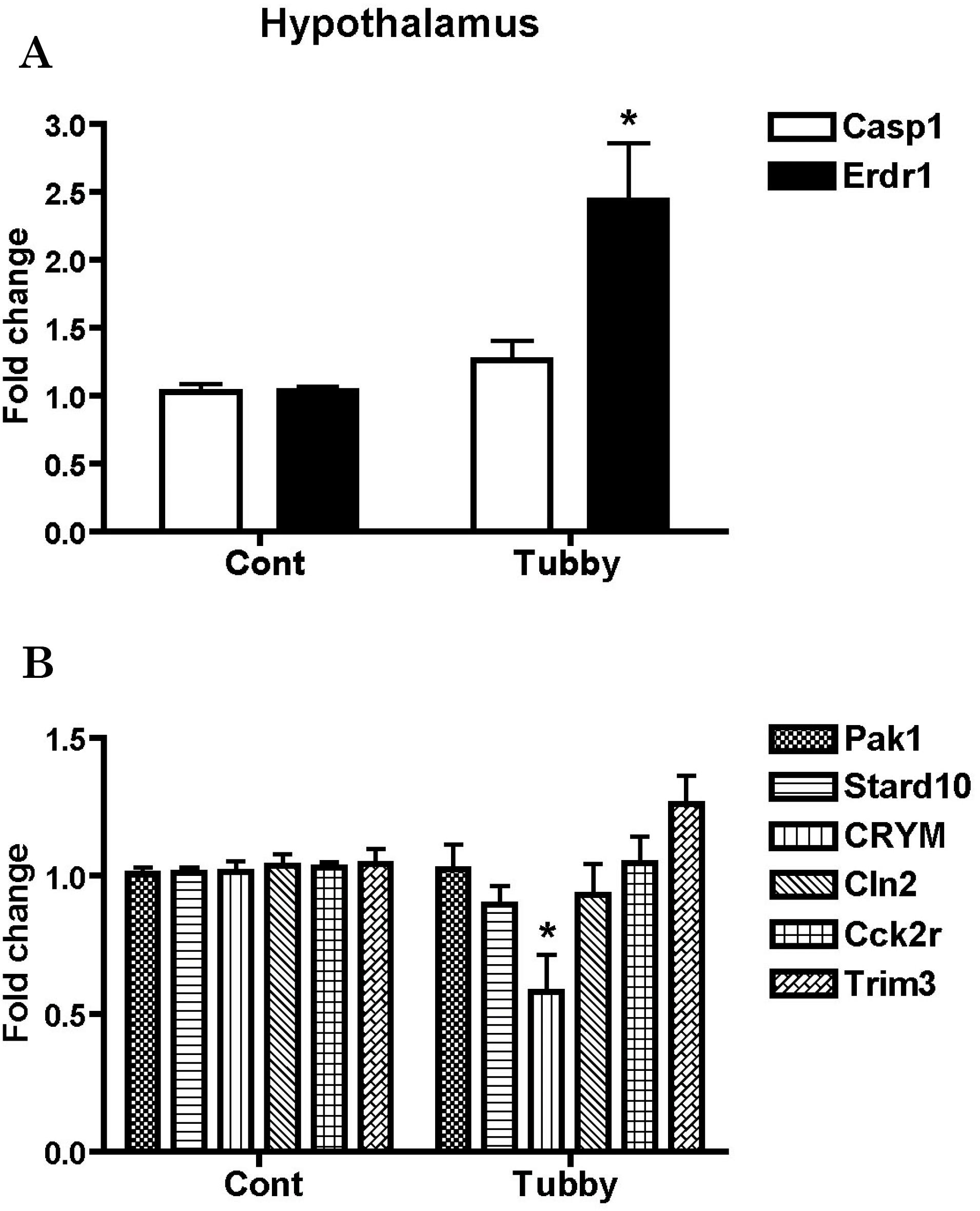 | Fig. 2.Validation of array-based gene expression profiles in the hypothalamus of tubby mice using real-time RT-PCR. mRNA levels of genes were determined using real-time RT-PCR. Hypothalamic Casp1 and Erdr1 expressions (A), the up-regulated genes in the cortex, and the expression profiles of other down-regulated genes in the hypothalamus (B) are represented as fold changes compared to the hypothalmus of control mice. Expression levels of β-actin were used to normalize the values. ∗p<0.05. |
Table 1.
Sequences of primers used in real-time RT-PCR
Table 2.
A selection of genes up- or down-regulated in the cerebral cortex of tubby mice compared to control mice. Gene profiling was performed using microarray analysis. Expression level is presented as the mean of fold change (n=3). Negative fold change values indicate decreased gene expression in tubby mice
Table 3.
A selection of genes up- or down-regulated in the hypothalamus of tubby mice compared to control mice. Gene profiling was performed using microarray analysis. Expression level is presented as the mean of fold change (n=3). Negative fold change values indicate decreased gene expression in tubby mice




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download