Abstract
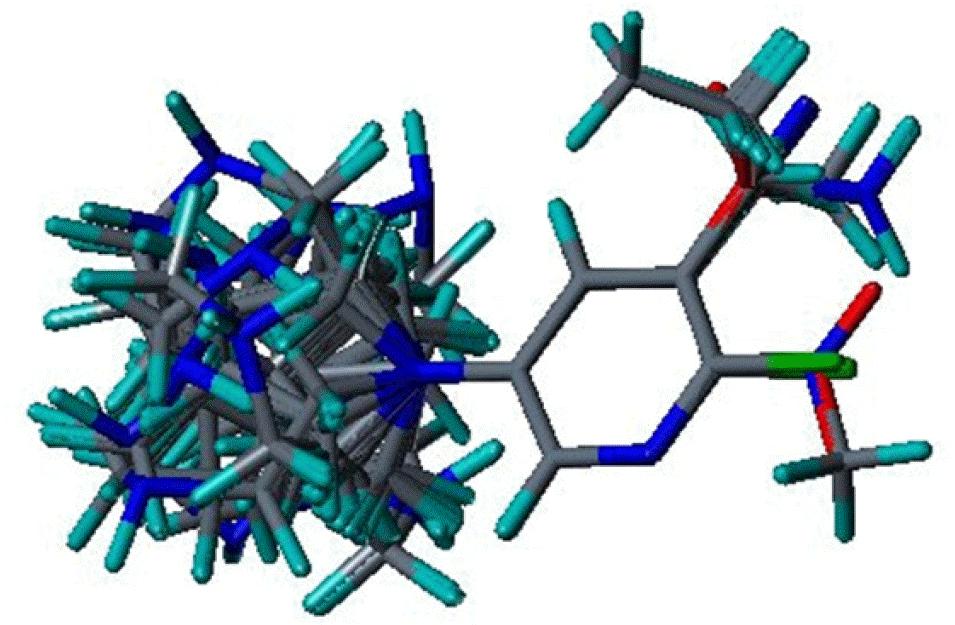
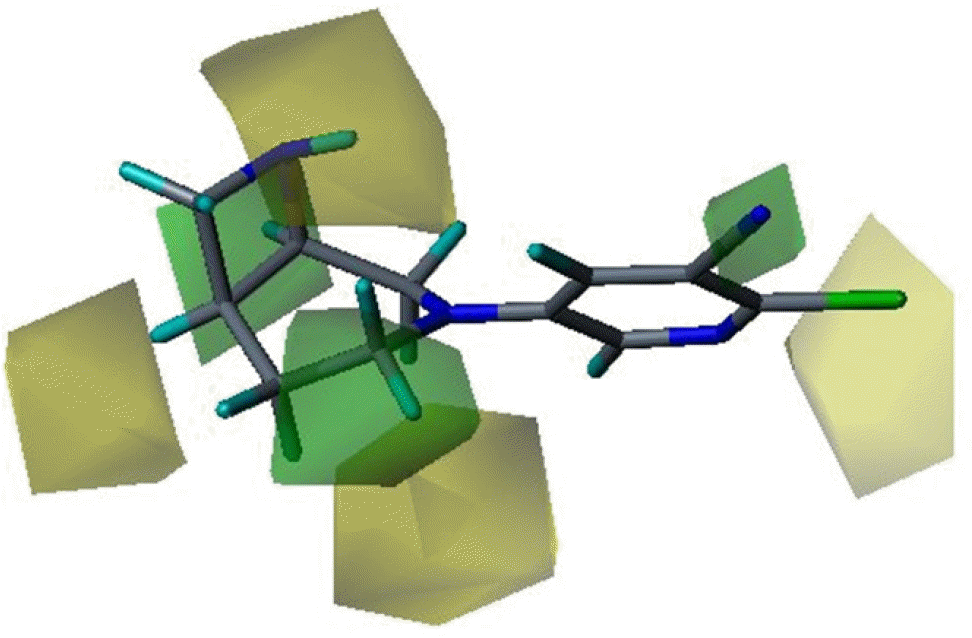
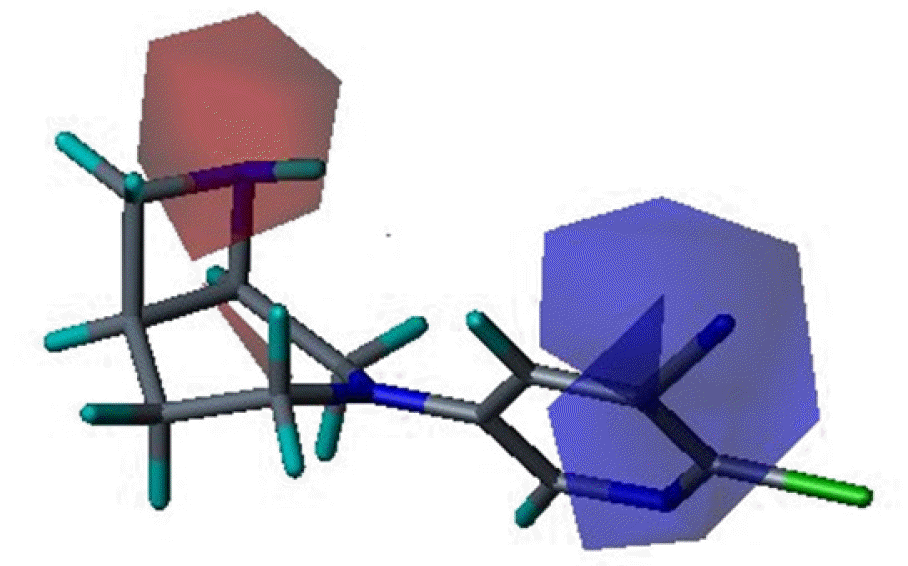
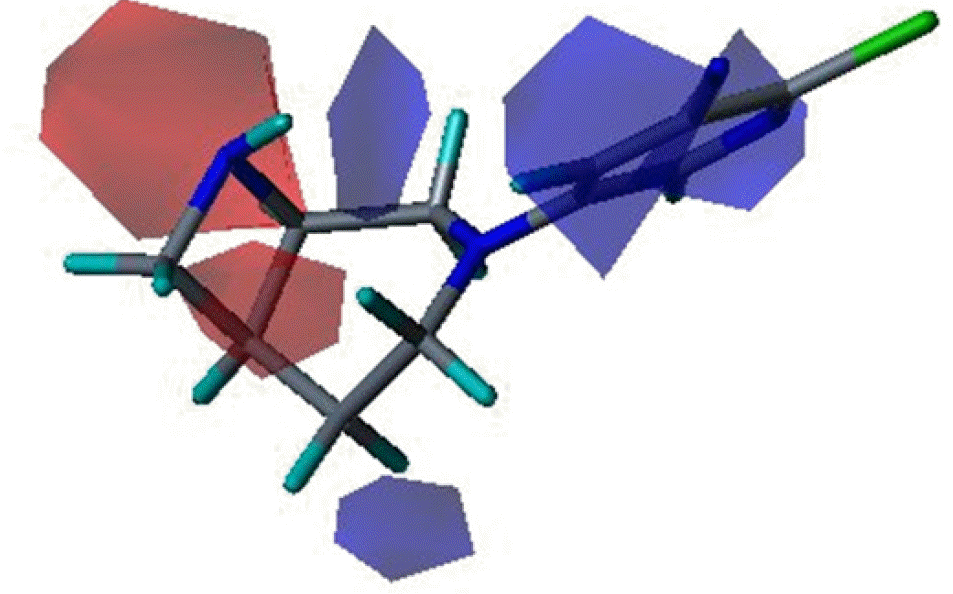
Three dimensional quantitative structure activity relationship between diazabicyclo[4.2.0]octanes and nicotinic acetylcholine receptor (hα4β2 and hα3β4) agonists was studied using comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA). From 11 CoMFA and CoMSIA models, CoMSIA with steric and electrostatic fields gave the best predictive models (q2=0.926 and 0.945, r2ncv=0.983 and 0.988). This study can be used to develop potent hα4β2 receptor agonists with low activity on hα3β4 subtype.
Go to : 
REFERENCES
Cashin AL., Torrice MM., McMenimen KA., Lester HA., Dougherty DA. Chemical-scale studies on the role of a conserved aspartate in preorganizing the agonist binding site of the nicotinic acetylcholine receptor. Biochemistry. 46:630–639. 2007.

Dehkordi O., Millis RM., Dennis GC., Jazini E., Williams C., Hussain D., Jayam-Trouth A. Expression of α-7 and α-4 nicotinic acetylcholine receptors by GABAergic neurons of rostral ventral medulla and caudal pons. Brain Res. 1185:95–102. 2007.
Dougherty JJ., Wu J., Nichols RA. β-Amyloid regulation of presynaptic nicotinic receptors in rat hippocampus and neocortex. J Neurosci. 23:6740–6747. 2003.

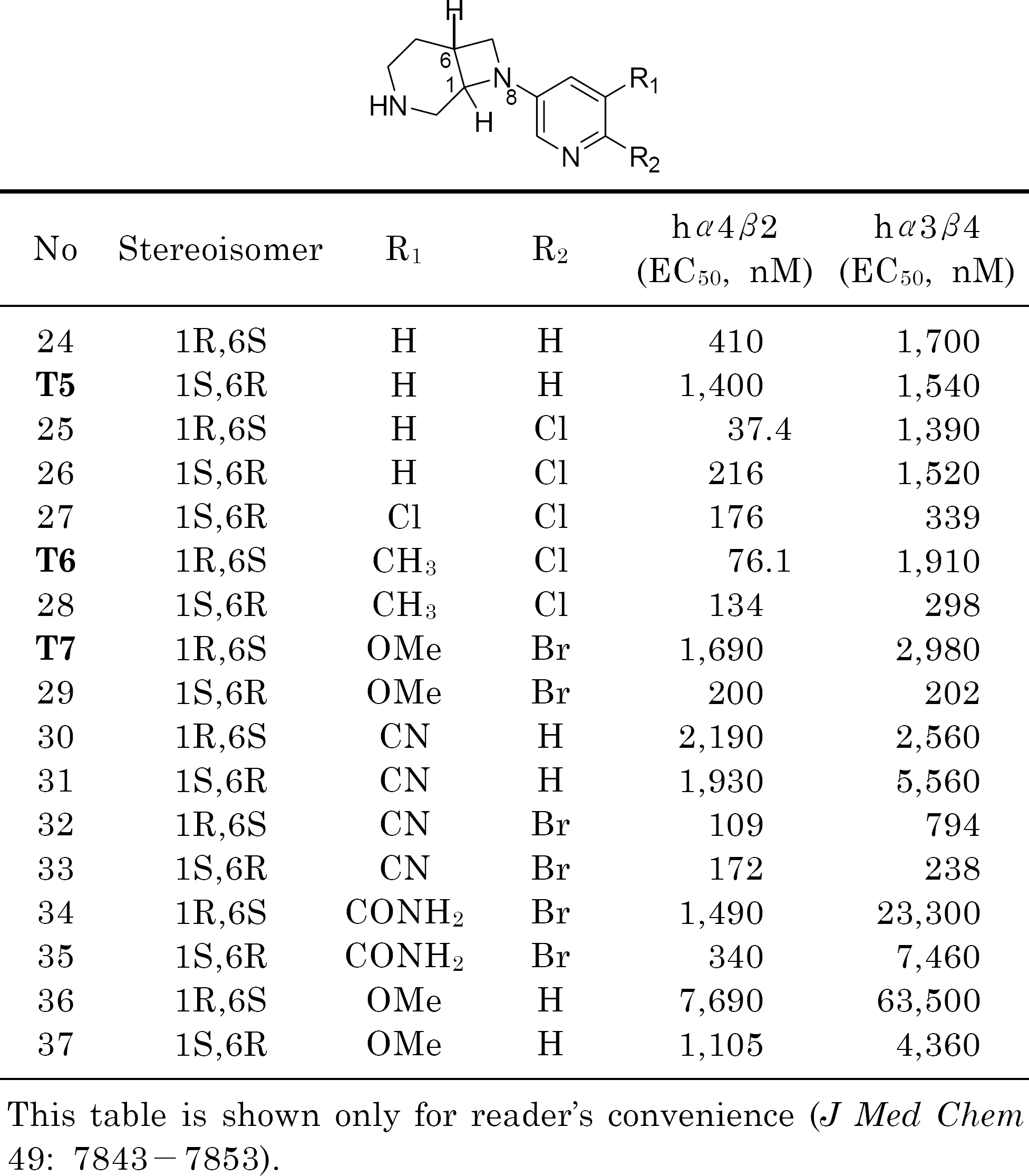
Frost MJ., Bunnelle HW., Tietje KR., Anderson DJ., Rueter LE., Curzon P., Surowy CS., Jerome JJ., Kohlhaas DK., Buckley MJ., Henry RF., Dyhring T., Ahring PK., Meyer MD. Synthesis and structure-activity relationships of 3,8-diazabicyclo[4.2.0]octane ligands, potent nicotinic acetylcholine receptor agonists. J Med Chem. 49:7843–7853. 2006.
Girod R., Barazangi N., McGehee D., Role LW. Facilitation of glutamatergic neurotransmission by presynaptic nicotinic acetylcholine receptors. Neuropharmacology. 39:2715–2725. 2000.

Gotti C., Zoli M., Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance Trends. Pharmacol Sci. 27:482–491. 2006.
Grady SR., Salminen O., Laverty DC., Whiteaker P., McIntosh JM., Collins AC., Marks MJ. The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochem Pharmacol. 74:1235–1246. 2007.

Hays JT., Ebbert J., Sood A. Efficacy and safety of varenicline for smoking cessation. Am J Med. 121:S32–S42. 2008.

Hogg RC., Bertrand D. Neuroscience: What genes tell us about nicotine addiction. Science. 306:983–985. 2004.
Jensen AA., Fr⊘lund B., Liljefors T., Krogsgaard LP. Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J Med Chem. 48:4705–4745. 2005.

Kenny PJ., File SE., Neal MJ. Evidence for a complex influence of nicotinic acetylcholine receptors on hippocampal serotonin release. J Neurochem. 75:2409–2414. 2000.

Kuryatov A., Onksen J., Lindstrom J. Roles of accessory subunits in α4β2 nicotinic receptors. Molecular Pharmacology. 74:132–143. 2008.

Lape R., Colquhoun D., Sivilotti LG. On the nature of partial agonism in the nicotinic receptor superfamily. Nature. 454:722–727. 2008.

O'Leary KT., Parameswaran N., Johnston LC., McIntosh JM., Di Monte DA., Quik M. Paraquat exposure reduces nicotinic receptor-evoked dopamine release in monkey striatum. J Pharmacol Exp Ther. 327:124–129. 2008.
Owen RT., Serradell N., Rosa E. Ispronicline: nicotinic acetylcholine α4β2 agonist treatment of cognition disorders. Drugs of the Future. 33:197–202. 2008.
Pons S., Fattore L., Cossu G., Tolu S., Porcu E., McIntosh JM., Changeux JP., Maskos U., Fratta W. Crucial role of α4 and α6 nicotinic acetylcholine receptor subunits from ventral tegmental area in systemic nicotine self-administration. J Neurosci. 28:12318–12327. 2008.

Vincler M., McIntosh JM. Targeting the α9 α10 nicotinic acetylcholine receptor to treat severe pain. Expert Opinion on Therapeutic Targets. 11:891–897. 2007.
SYBYL Molecular Modeling Software. Tripos Inc.;St. Louis, USA: 2008.
Go to : 
Table 3.
CoMFA and CoMSIA results of the training set
| Field∗ | q2† | N‡ | SEP§ | r2ncv|| | SEE¶ | F∗∗ | Contributions | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S | E | H | D | A | |||||||
| hα4β2 subtype | |||||||||||
| CoMFA | |||||||||||
| S | 0.832 | 6 | 0.375 | 0.967 | 0.166 | 147.107 | 1 | ||||
| E | 0.819 | 6 | 0.390 | 0.966 | 0.168 | 143.232 | 1 | ||||
| SE | 0.892 | 6 | 0.301 | 0.987 | 0.104 | 382.064 | 0.534 | 0.466 | |||
| CoMSIA | |||||||||||
| SE | 0.926 | 6 | 0.249 | 0.983 | 0.120 | 285.165 | 0.128 | 0.872 | |||
| SEH | 0.695 | 6 | 0.506 | 0.987 | 0.103 | 393.914 | 0.077 | 0.567 | 0.356 | ||
| SED | 0.866 | 6 | 0.335 | 0.978 | 0.134 | 227.120 | 0.056 | 0.454 | 0.490 | ||
| SEA | 0.884 | 6 | 0.312 | 0.978 | 0.136 | 220.764 | 0.117 | 0.782 | 0.101 | ||
| SEDA | 0.833 | 6 | 0.374 | 0.973 | 0.151 | 178.867 | 0.048 | 0.395 | 0.506 | 0.050 | |
| SEHD | 0.595 | 1 | 0.608 | 0.980 | 0.129 | 248.687 | 0.046 | 0.339 | 0.199 | 0.415 | |
| SEHA | 0.650 | 6 | 0.542 | 0.986 | 0.110 | 344.311 | 0.067 | 0.422 | 0.356 | 0.156 | |
| SEHDA | 0.605 | 5 | 0.576 | 0.987 | 0.104 | 386.129 | 0.034 | 0.249 | 0.226 | 0.363 | 0.127 |
| hα3β4 subtype | |||||||||||
| CoMFA | |||||||||||
| S | 0.787 | 6 | 0.518 | 0.948 | 0.255 | 91.574 | 1 | ||||
| E | 0.830 | 6 | 0.462 | 0.969 | 0.197 | 157.454 | 1 | ||||
| SE | 0.934 | 6 | 0.288 | 0.985 | 0.139 | 319.679 | 0.512 | 0.488 | |||
| CoMSIA | |||||||||||
| SE | 0.945 | 6 | 0.262 | 0.988 | 0.125 | 399.560 | 0.152 | 0.848 | |||
| SEH | 0.688 | 6 | 0.627 | 0.979 | 0.162 | 234.901 | 0.084 | 0.529 | 0.387 | ||
| SED | 0.900 | 6 | 0.354 | 0.975 | 0.179 | 191.750 | 0.061 | 0.426 | 0.513 | ||
| SEA | 0.871 | 6 | 0.402 | 0.976 | 0.174 | 202.120 | 0.148 | 0.773 | 0.079 | ||
| SEDA | 0.868 | 5 | 0.408 | 0.972 | 0.189 | 171.724 | 0.057 | 0.404 | 0.484 | 0.055 | |
| SEHD | 0.717 | 4 | 0.600 | 0.981 | 0.154 | 259.610 | 0.038 | 0.306 | 0.204 | 0.452 | |
| SEHA | 0.669 | 6 | 0.645 | 0.975 | 0.176 | 197.341 | 0.074 | 0.368 | 0.367 | 0.190 | |
| SEHDA | 0.749 | 4 | 0.571 | 0.983 | 0.147 | 287.861 | 0.034 | 0.216 | 0.232 | 0.388 | 0.129 |
Table 4.
CoMSIA actual and predicted activity (pEC50) of the training set
Table 5.
CoMSIA actual and predicted activity (pEC50) of the test set




 PDF
PDF ePub
ePub Citation
Citation Print
Print


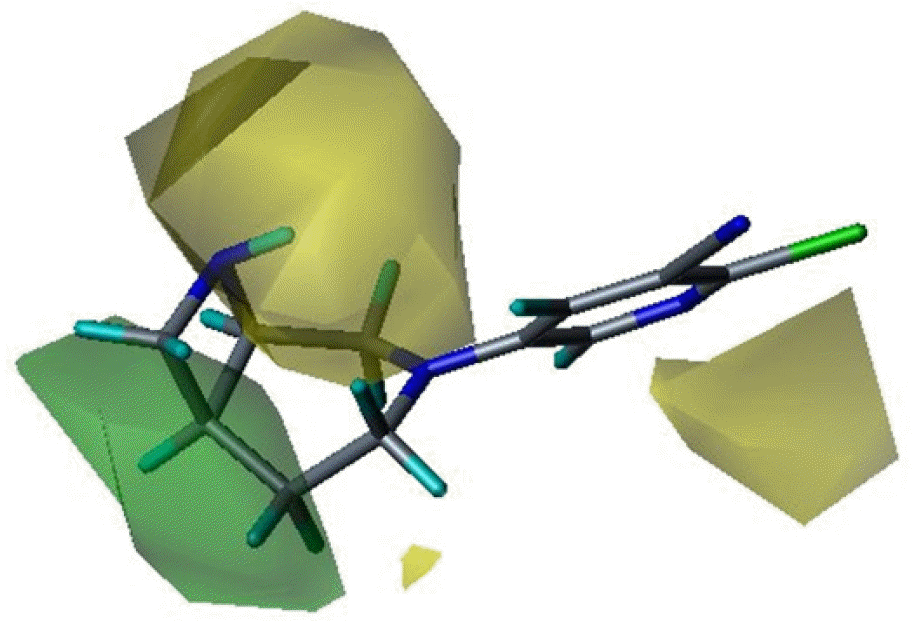
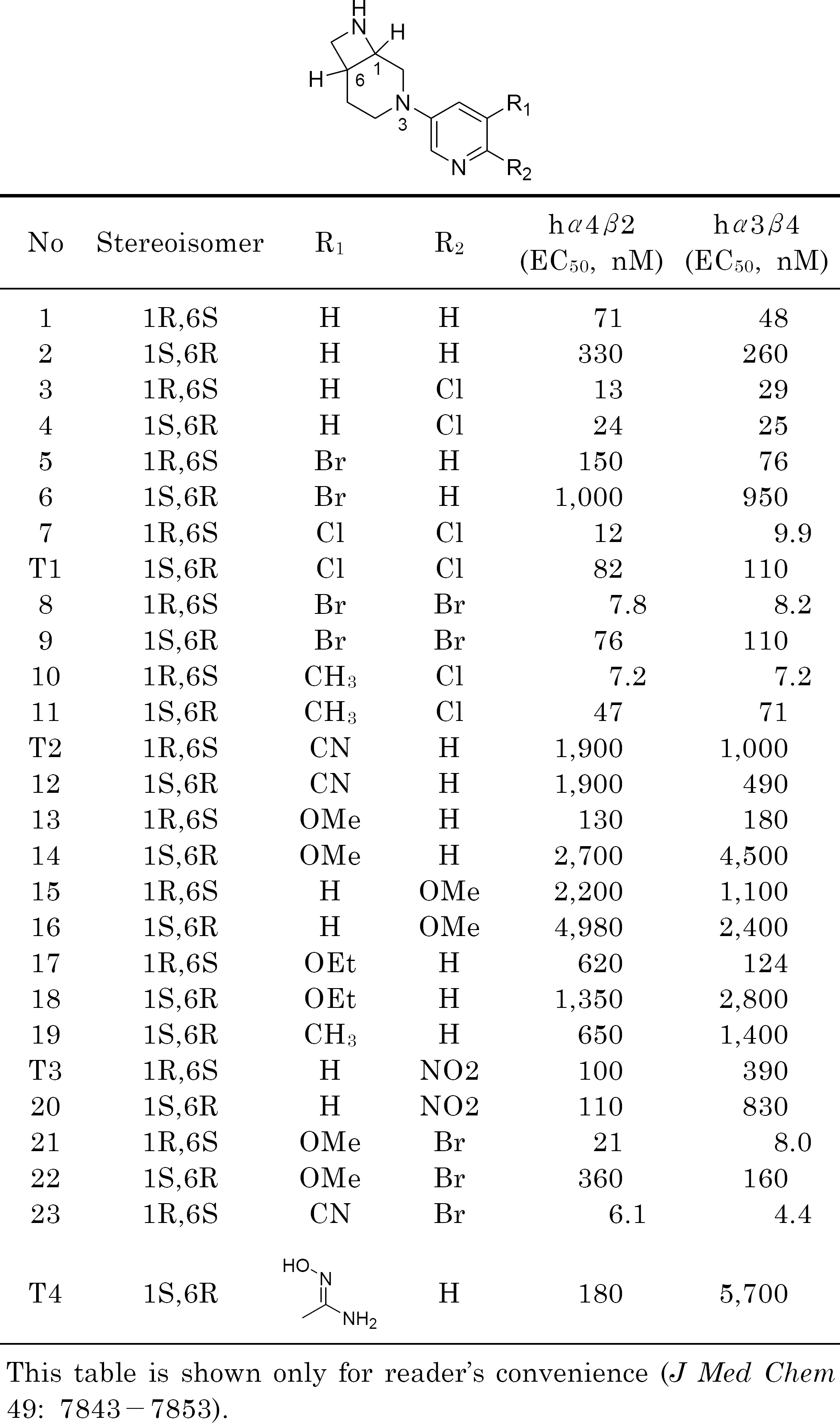
 XML Download
XML Download