Lung cancer is the most common cause of cancer-related deaths. Although the prognosis of lung cancer has improved in the last two decades, the 5-year survival rate is still poor (approximately 17%) (
1). Non-small cell lung cancer (NSCLC), which makes up about 85% of primary lung cancers, is potentially curable by surgical resection in the early cancer stage. However, 30% to 55% of NSCLC patients who underwent curative surgical resection eventually developed cancer recurrence and died of their disease (
2). Therefore, many researchers are trying to find predictive markers for recurrence or prognostic markers for survival outcome.
The WNT signaling pathway is a stem cell pathway that has important roles in embryonic development and tissue regeneration (
3,
4). This signaling pathway was reported to be associated with carcinogenesis in many tissues (
5). After the discovery of an oncogenic effect of
WNT1 in a mouse model, the association between the WNT signaling pathway and human cancer has been studied actively (
678910). The WNT signaling pathway has also been reported to affect the pathogenesis of NSCLC (
11,
12). Overexpression of the
WNT gene in NSCLC is thought to be associated with poor prognosis (
13,
14). Recently, Coscio et al. (
15) investigated the association between single nucleotide polymorphisms (SNPs) of WNT signaling pathway genes and the prognosis of NSCLC in Caucasians, and they reported that several SNPs of the WNT pathway were associated with cancer recurrence and survival of patients with early stage NSCLC.
The effect of genetic variants on survival outcome may be different depending on the ethnic group. Therefore, we investigated whether the SNPs of WNT signaling pathway genes identified in Caucasians had the same associations in patients with stages I–IIIA NSCLC in a Korean population.
A total of 761 patients were enrolled. Of these, 354 patients who had been diagnosed with stage I, II or IIIA NSCLC and underwent curative surgical resection at the Kyungpook National University Hospital (KNUH) from September 1998 to August 2007; 407 patients with surgically resected NSCLC for curative purpose collected by Seoul National University Hospital (SNUH) between September 2005 and October 2010 were also included in this study. All of the patients in this study were ethnic Koreans. Blood samples for genotyping were collected before surgery. The patients who received neoadjuvant chemotherapy were excluded, to avoid confounding effects on the DNA. Blood samples were provided by the National Biobank of Korea, which is supported by the Ministry of Health, Welfare, and Family Affairs. Written informed consent was obtained from all study participants. This study was approved by the Institutional Review Broad of Kyungpook National University of Hospital (Approval No., KNUMCBIO_11-0001).
Seven SNPs (rs4135385, rs10898563, rs503022, rs629537, rs11658976, rs3765351, and rs713065), which were associated with overall survival after adjustment for multiple testing (
q < 0.1) in the study of Coscio et al. (
15), were selected. The rs2536182, which was the most significantly associated with recurrence-free survival and validated in patients from Mayo Clinic, was also selected (
15). A total of eight SNPs were genotyped using SEQUENOM’s MassARRAY
® iPLEX assay (SEQUENOM Inc., San Diego, CA, USA). For quality control, the genotyping analysis was performed blind with regard to the subjects.
Continuous variables were compared by Student’s t test, and categorical variables were examined using the Chi-square test. OS was counted from the day of surgery to the date of death or the last follow-up. Disease-free survival (DFS) was measured from the day of surgery until recurrence or death from any cause. The Kaplan-Meier method and log-rank tests were used to compare OS and DFS according to genotype. To estimate hazard ratios and 95% confidence intervals (CIs), multivariate Cox proportional analysis was used with adjustment for age (< 65 years vs. ≥ 65 years), gender (male vs. female), smoking status (smoker vs. non-smoker), histological type (squamous cell carcinoma vs. adenocarcinoma), pathological stage (stage I vs. stage II or IIIA), and adjuvant therapy (yes vs. no). A P value of less 0.05 was considered statistically significant. All analyses were performed using Statistical Analysis System for Windows, version 9.2 (SAS Institute, Cary, NC, USA).
Demographic and clinical characteristics of the patients are summarized in
Supplementary Table 1. Among the 761 patients, 206 (29.1%) deaths occurred and the estimated 5-year OS was 62% (95% CI, 57%–67%). Upon univariate analysis, the pathological stage was found to be significantly associated with OS (Log-Rank
P [
P
L-R] < 1 × 10
-4). Age, gender, and smoking status were also associated with OS. The estimated 5-year DFS was 45% (95% CI, 40%–49%) and only the pathological stage was significantly associated with DFS (
P
L-R < 1 × 10
-4). The genotype frequencies of the eight SNPs were in Hardy-Weinberg equilibrium. The eight SNPs were not significantly associated with patient- or tumor related factors, such as age, gender, smoking status, histological type, or pathological stage (data not shown). Among 8 SNPs, the
WNT5A rs503022 and rs629537 were in strong linkage disequilibrium (LD) (
Supplementary Fig. 1).
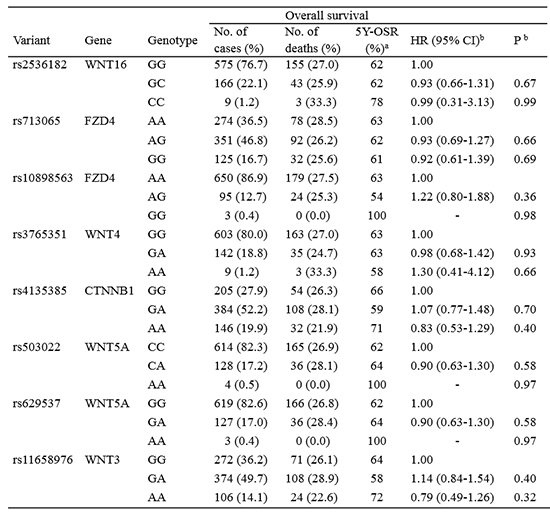
The associations between the eight SNPs of WNT signaling pathway genes and survival outcome of patients with NSCLC are shown in
Table 1. None of eight SNPs were significantly associated with OS or DFS. In addition, when patients were categorized according to age, gender, smoking status, and histological type, no significant associations were found between the SNPs and survival outcome (data not shown).
We attempted to determine the impact of SNPs of WNT signaling pathway genes on the survival outcome of Korean patients with NSCLC. However, this study found no significant association in this regard. In addition, there was no evidence of any effect modification by age, gender, smoking history, or tumor histology.
Coscio et al. (
15) reported that
CTNNB1 rs4135385,
FZD4 rs10898563,
WNT5A rs503022,
WNT5A rs629537,
WNT3 rs11658976, and
WNT4 rs3765351 were associated with poor OS in stage I or II NSCLCs. The
FZD4 rs713065 variant was associated with better OS (
15). In this study, the SNPs related with OS in the study of Coscio et al. (
15) were not associated with survival outcome in Korean NSCLC patients. In addition, there were no differences in OS according to SNPs in patients with stage I or II NSCLC (
Supplementary Table 2).
Although the reason for the discrepancy in the two studies is unclear, the different genetic backgrounds of Caucasians and Koreans may be a major reason. For example, the minor allele frequency of rs10898563 is 0.35 in Caucasians but 0.07 in Koreans (
Supplementary Table 2). Differences in minor allele frequencies according to ethnic groups can dramatically reduce the statistical power of a replication study (
16). The heterogeneity of LD patterns across populations could be another reason for the replication failure (
17). Because true functional variant(s) may be linked to the investigated variants, differences in LD patterns across ethnic groups can be a confounding factor for a replication study. Therefore, additional studies are needed to clarify the effect of WNT signaling pathway gene SNPs on the survival outcome of NSCLCs in diverse ethnic groups. Further studies of other SNPs in the study of Coscio et al. (
15) are also needed.
In conclusion, the present study found that SNPs of WNT signaling pathway genes, which were related to survival outcome in Caucasian NSCLC patients, did not have the same significant association in Korean NSCLC patients.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download