INTRODUCTION
MATERIALS AND METHODS
Cell culture
Fluorescence activated cell sorting (FACS)
Drug treatment
RNA isolation and semi-quantitative RT-PCR
Table 1
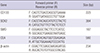
RT-PCR Primers used in this study
Cell viability assay
Immunofluorescence analysis (IF)
Sphere formation assay
In vivo xenograft tumorigenecity assay
RESULTS
Stemness properties of SW620 CD133+ cells
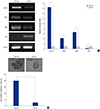 | Fig. 1Stemness properties of sorted SW620 CD133+ cells. (A) The mRNA expression of stemness and differentiation marker in SW620 CD133+ and CD133–cells were measured by RT-PCR. β-Actin was used as a loading control. For comparisons, the relative value for markers of CD133–cells was considered to be “1”. (B) Self-renewal ability of sorted cells was analyzed by sphere formation assay. Pictures were taken at ×40 magnification. Scale bar = 100 µm. Data are expressed as the mean ± standard error of the mean (SEM) of three independent experiments performed (*P < 0.001). |
Differentiation induction of SW620 CD133+ cells by dual-inhibition of PI3K and mTOR
 | Fig. 2Stemness and differentation properties of PI3K and/or mTOR inhibitors treated SW620 CD133+ cells. (A) The mRNA expression of stemness and differentiation markers after inhibition of PI3K and/or mTOR was measured by RT-PCR. β-Actin was used as a loading control. For comparisons, relative intensity for markers of DMSO-treated CD133+ cells was defined as “1”. *P < 0.001. (B) The protein expression of CD133 was measured by Immunofluorescence assay (IF). CD133 was stained in red. (C) The protein expression of CEA was measured by IF. CEA were stained in green, respectively. In each experiment (B and C), the nuclei were counterstained with DAPI (blue). For comparisons, relative intensity of Corrected Total Cell Fluorescence (CTCF) for CD133 and CEA of DMSO-treated CD133+ cells was defined as “1”. Data are expressed as the mean ± standard error of the mean (SEM) of three independent experiments performed (*P < 0.001, “N/S” means “statistically not significant”). Pictures were taken at ×400 magnification. Scale bar = 10 μm. |
Drug sensitivity change in differentiated SW620 CD133+ cells by dual-inhibition
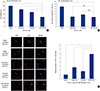 | Fig. 3The drug sensitivity of inhibited SW620 CD133+ cells. (A) Inhibited SW620 CD133+ cells were treated with paclitaxel (100 nM) for 24 hours and cell viability was assessed by CCK-8 assay. For comparisons cell viability of DMSO treated CD133+ cells was 100%. (B) Self-renewal ability of inhibited SW620 CD133+ cells treated with paclitaxel and self-renewal ability was assessed by sphere formation assay. (C) Differentiation marker (CEA) expression of inhibited CSCs treated with paclitaxel. The protein expression of CEA was assessed by IF. For the comparison, relative intensity of CTCF for CEA of DMSO treated CD133+ cells was defined as “1”. All data are expressed as the mean ± standard error of the mean (SEM) of three independent experiments performed (*P < 0.05, †P < 0.01, ‡P < 0.001, “N/S” means “statistically not significant”). Pictures were taken at ×400 magnification. Scale bar = 10 μm. |
Tumor growth and the sensitivity to the anti-cancer drug by dual-blocking of PI3K and mTOR
 | Fig. 4Xenograft tumorigenecity assay of inhibitors treated SW620 CD133+ cells before and after injection of anti-cancer drug. (A) Comparison of tumorigenecity of inhibitor treated SW620 CD133+ cells in BALB/c nude mice. Balb/c nude mice were subcutaneously injected with sorted SW620 CD133+ cells. The left flank was injected with untreated CD133+ cells, while the right flank was injected with CD133+ cells treated with each inhibitor. Anti-cancer drug (paclitaxel) was injected since 28 days. (B) Tumor volume (mm3) of xenografts from Balb/c mice. The change in tumor volume was checked for each group. DMSO treated group was used as a control. Data are expressed as the mean ± standard error of the mean (SEM) of mice for each group. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download