INTRODUCTION
MATERIALS AND METHODS
Cell culture
Patients and tissue samples
Immunohistochemical analysis and evaluation of eIF3a expression
RNA isolation and semi-quantitative reverse transcription–PCR
Western blot analysis
Cell viability detection
Colony formation assay
Wound-healing assay
Transwell assay
Mouse xenografts
Statistical analysis
RESULTS
Aberrant eIF3a expression in human pancreatic cancer tissues and the eIF3a expression correlated with tumor metastasis and tumor staging
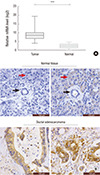 | Fig. 1Aberrant eIF3a expression in pancreatic cancer tissues.
(A) Relative eIF3a mRNA levels in the pancreatic ductal adenocarcinoma tissues (n = 30) and their paired adjacent non-cancerous pancreas tissues (n = 30). (B) Immunohistochemistry analysis of the protein level of eIF3a in slides from normal pancreas tissues and the pancreatic ductal adenocarcinoma tissues (n = 140). It was shown that eIF3a was non-expressed or lowly expressed in normal pancreatic duct (black arrow) and normal pancreatic acini (red arrow). In contrast, eIF3a was strongly expressed in the cancer tissues.
*P < 0.001
|
Table 1
Association between eIF3a expression and the clinicopathological variables in 140 cases of pancreatic ductal adenocarcinoma
Constitutive eIF3a expression in human pancreatic cancer cell lines and knockdown of its expression in vitro
 | Fig. 2The constitutive expression of eIF3a in pancreatic cancer cell lines and the knockdown efficacy of a specific shRNA against eIF3a. (A) In the seven pancreatic cancer cell lines, it was observed that SW1990 and Capan-1 cells exhibited the strongest expression of eIF3a, whereas Miapaca-2 cell line exhibited the least expression of eIF3a. Hence, SW1990 and Capan-1 were chosen for subsequent analyses. (B) A specific shRNA against eIF3a was utilized to knock down the expression of eIF3a in SW1990 cells and Capan-1 cells. Western blot analysis revealed that the protein level of eIF3a was barely detected after transfection of the specific shRNA into these two cell lines, suggesting the high efficacy of our designed shRNA. |
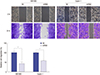
Knockdown of eIF3a inhibited cell proliferation and colony formation in pancreatic cancer cells
 | Fig. 3Knockdown of eIF3a inhibited cell proliferation and colony formation in pancreatic cancer cell lines. (A) In the cell proliferation assay, significant disparities were observed from the fourth day in SW1990 cells and the fifth day in Capan-1 cells. The inhibitory effects grew as the time extended in both cells. (B) In the colony formation assay, it was shown that knockdown of eIF3a caused visual decreases of colonies in both cell lines. (C) By counting the colonies numbers, it was further shown that more than 60% of SW1990 colonies and 75% of Capan-1 colonies were suppressed upon eIF3a shRNA transfection. Data were obtained in triplicate with each experiments repeated three times.
*P < 0.05; †P < 0.01
|
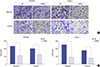
Knockdown of eIF3a inhibited wound recovery abilities in pancreatic cancer cells
 | Fig. 4Knockdown of eIF3a inhibited the wound recovery in SW1990 cells and Capan-1 cells. Both SW1990 cells and Capan-1 cells were subject to wound-healing assays after transfection with scramble or specific shRNA against eIF3a. Twenty-four hours after the scratch, wound recovery rates were photographed and the recovered areas which represented cell migration were quantified and averaged from three independent assays.
*P < 0.01
|
Knockdown of eIF3a decreased cell migration and invasion abilities in human pancreatic cancer cell lines
 | Fig. 5Knockdown of eIF3a decreased cell migration and invasion abilities in human pancreatic cancer cell lines. (A) SW1990 cells and Capan-1 cells were subject to transwell assays after cells were depleted of eIF3a by shRNA. Visually, the transmigrated cells were significantly decreased in the shRNA group relative to control group. (B) After counting the transmigrated cells, it was shown that the migration rate was inhibited by 55% for SW1990 cells and 46% for Capan-1 cells. Meanwhile, cell invasion rate was decreased by more than 60% for both cell lines. Each assay was repeated for three times.
*P < 0.01
|
Knockdown of eIF3a inhibited the tumorigenic ability in a xenotransplanted model
 | Fig. 6Knockdown of eIF3a inhibited the tumorigenic ability in a xenotransplanted model. (A) Capan-1 cells with or without eIF3a depletion were subcutaneously injected into the nude mice (n = 5 for each group) to determine the impact of eIF3a depletion on pancreatic tumor growth in vivo. Tumor volumes from two groups of mice were monitored for a consecutive 4 weeks. (B) After four weeks, tumors were dissected. (C) Tumor weights from each group were weighed. It was shown that the average tumor weight from eIF3a-depleted group was significantly decreased as compared with that in the control group.
*P < 0.05
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download