1. Shwayder T. Ichthyosis in a nutshell. Pediatr Rev. 1999; 20:5–12.
2. Cañueto J, Ciria S, Hernández-Martín A, Unamuno P, González-Sarmiento R. Analysis of the STS gene in 40 patients with recessive X-linked ichthyosis: a high frequency of partial deletions in a Spanish population. J Eur Acad Dermatol Venereol. 2010; 24:1226–1229.
3. Sugawara T, Nomura E, Hoshi N. Both N-terminal and C-terminal regions of steroid sulfatase are important for enzyme activity. J Endocrinol. 2006; 188:365–374.
4. Pehlke JR, Venkataramani V, Emmert S, Mohr A, Zoll B, Nau R. X-linked recessive ichthyosis (XRI), cerebellar ataxia and neuropsychiatric symptoms. Fortschr Neurol Psychiatr. 2013; 81:40–43.
5. Macsai MS, Doshi H. Clinical pathologic correlation of superficial corneal opacities in X-linked ichthyosis. Am J Ophthalmol. 1994; 118:477–484.
6. Crowe MA, James WD. X-linked ichthyosis. JAMA. 1993; 270:2265–2266.
7. Traupe H, Müller D, Atherton D, Kalter DC, Cremers FP, van Oost BA, Ropers HH. Exclusion mapping of the X-linked dominant chondrodysplasia punctata/ichthyosis/cataract/short stature (Happle) syndrome: possible involvement of an unstable pre-mutation. Hum Genet. 1992; 89:659–665.
8. Fan X, Petruschka L, Wulff K, Grimm U, Herrmann FH. Biochemical and immunological characterization of X-linked ichthyosis. J Inherit Metab Dis. 1993; 16:17–26.
9. Li M. Gene deletion of X-linked ichthyosis. Zhonghua Yi Xue Za Zhi (Taipei). 1992; 72:210–212.
10. Newman RS, Affara NA, Yates JR, Mitchell M, Ferguson-Smith MA. Physical mapping of deletion breakpoints in patients with X-linked ichthyosis: evidence for clustering of distal and proximal breakpoints. Proc Biol Sci. 1990; 242:231–239.
11. Okano M, Yoshimoto K. Commercial assay for steroid sulphatase activity in X-linked ichthyosis. Br J Dermatol. 1990; 123:546.
12. Serizawa S, Nagai T, Ito M, Sato Y. Cholesterol sulphate levels in the hair and nails of patients with recessive X-linked ichthyosis. Clin Exp Dermatol. 1990; 15:13–15.
13. Elias PM, Williams ML, Choi EH, Feingold KR. Role of cholesterol sulfate in epidermal structure and function: lessons from X-linked ichthyosis. Biochim Biophys Acta. 2014; 1841:353–361.
14. Jensen PK, Herrmann FH, Hadlich J, Bolund L. Proliferation and differentiation of cultured epidermal cells from patients with X-linked ichthyosis and ichthyosis vulgaris. Acta Derm Venereol. 1990; 70:99–104.
15. Herrmann FH, Wirth B, Wulff K, Hadlich J, Voss M, Gillard EF, Kruse TA, Ferguson-Smith MA, Gal A. Gene diagnosis in X-linked ichthyosis. Arch Dermatol Res. 1989; 280:457–461.
16. Jang JE, Kook HI. Two cases of sex-linked ichthyosis improved by an oral aromatic retinoid (Ro 10-9359). Korean J Dermatol. 1982; 20:431–436.
17. Cho YW, Hong CK, Song KY, Ro BI, Chang CY. A case of X-linked ichthyosis. Korean J Dermatol. 1990; 28:368–372.
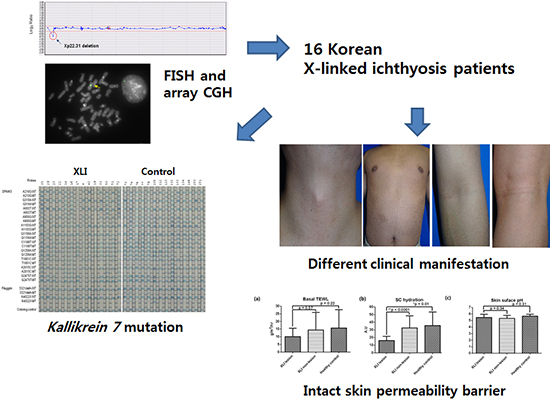
18. Park SJ, Jung EH, Ryu RS, Kang HW, Ko JM, Kim HJ, Cheon CK, Hwang SH, Kang HY. Clinical implementation of whole-genome array CGH as a first-tier test in 5080 pre and postnatal cases. Mol Cytogenet. 2011; 4:12.
19. Park SJ, Jung EH, Ryu RS, Kang HW, Chung HD, Kang HY. The clinical application of array CGH for the detection of chromosomal defects in 20,126 unselected newborns. Mol Cytogenet. 2013; 6:21.
20. Williams ML. Epidermal lipids and scaling diseases of the skin. Semin Dermatol. 1992; 11:169–175.
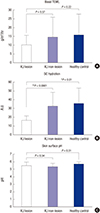
21. Lavrijsen AP, Oestmann E, Hermans J, Boddé HE, Vermeer BJ, Ponec M. Barrier function parameters in various keratinization disorders: transepidermal water loss and vascular response to hexyl nicotinate. Br J Dermatol. 1993; 129:547–554.
22. Johansen JD, Ramsing D, Vejlsgaard G, Agner T. Skin barrier properties in patients with recessive X-linked ichthyosis. Acta Derm Venereol. 1995; 75:202–204.
23. Hoppe T, Winge MC, Bradley M, Nordenskjöld M, Vahlquist A, Berne B, Törmä H. X-linked recessive ichthyosis: an impaired barrier function evokes limited gene responses before and after moisturizing treatments. Br J Dermatol. 2012; 167:514–522.
24. Groen D, Poole DS, Gooris GS, Bouwstra JA. Investigating the barrier function of skin lipid models with varying compositions. Eur J Pharm Biopharm. 2011; 79:334–342.
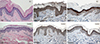
25. Elias PM, Crumrine D, Rassner U, Hachem JP, Menon GK, Man W, Choy MH, Leypoldt L, Feingold KR, Williams ML. Basis for abnormal desquamation and permeability barrier dysfunction in RXLI. J Invest Dermatol. 2004; 122:314–319.
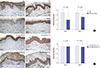
26. Ekholm IE, Brattsand M, Egelrud T. Stratum corneum tryptic enzyme in normal epidermis: a missing link in the desquamation process? J Invest Dermatol. 2000; 114:56–63.
27. Elias PM, Williams ML, Holleran WM, Jiang YJ, Schmuth M. Pathogenesis of permeability barrier abnormalities in the ichthyoses: inherited disorders of lipid metabolism. J Lipid Res. 2008; 49:697–714.
28. Jung M, Choi J, Lee SA, Kim H, Hwang J, Choi EH. Pyrrolidone carboxylic acid levels or caspase-14 expression in the corneocytes of lesional skin correlates with clinical severity, skin barrier function and lesional inflammation in atopic dermatitis. J Dermatol Sci. 2014; 76:231–239.
29. Trisnowati N, Soebono H, Sadewa AH, Kunisada M, Yogianti F, Nishigori C. A novel filaggrin gene mutation 7487delC in an Indonesian (Javanese) patient with atopic dermatitis. Int J Dermatol. 2016; 55:695–697.
30. Kang TW, Lee JS, Oh SW, Kim SC. Filaggrin mutation c.3321delA in a Korean patient with ichthyosis vulgaris and atopic dermatitis. Dermatology. 2009; 218:186–187.
31. Seo SJ, Park MK, Jeong MS, Kim JY, Kim IS. Clinical characteristics correlates with impaired barrier in filaggrin gene related Korean atopic dermatitis patients. In : 72nd Society for Investigative Dermatology (SID) Annual Meeting; 2012 May 9-12; Raleigh, North Carolina. Cleveland (OH): Society for Investigative Dermatology;2012.
32. Li M, Liu Q, Liu J, Cheng R, Zhang H, Xue H, Bao Y, Yao Z. Mutations analysis in filaggrin gene in northern China patients with atopic dermatitis. J Eur Acad Dermatol Venereol. 2013; 27:169–174.
33. Nomura T, Akiyama M, Sandilands A, Nemoto-Hasebe I, Sakai K, Nagasaki A, Ota M, Hata H, Evans AT, Palmer CN, et al. Specific filaggrin mutations cause ichthyosis vulgaris and are significantly associated with atopic dermatitis in Japan. J Invest Dermatol. 2008; 128:1436–1441.
34. Vasilopoulos Y, Cork MJ, Murphy R, Williams HC, Robinson DA, Duff GW, Ward SJ, Tazi-Ahnini R. Genetic association between an AACC insertion in the 3'UTR of the stratum corneum chymotryptic enzyme gene and atopic dermatitis. J Invest Dermatol. 2004; 123:62–66.
35. Hubiche T, Ged C, Benard A, Léauté-Labrèze C, McElreavey K, de Verneuil H, Taïeb A, Boralevi F. Analysis of SPINK 5, KLK 7 and FLG genotypes in a French atopic dermatitis cohort. Acta Derm Venereol. 2007; 87:499–505.
36. Hansson L, Bäckman A, Ny A, Edlund M, Ekholm E, Ekstrand Hammarström B, Törnell J, Wallbrandt P, Wennbo H, Egelrud T. Epidermal overexpression of stratum corneum chymotryptic enzyme in mice: a model for chronic itchy dermatitis. J Invest Dermatol. 2002; 118:444–449.
37. Kishibe M, Bando Y, Tanaka T, Ishida-Yamamoto A, Iizuka H, Yoshida S. Kallikrein-related peptidase 8-dependent skin wound healing is associated with upregulation of kallikrein-related peptidase 6 and PAR2. J Invest Dermatol. 2012; 132:1717–1724.
38. Hachem JP, Houben E, Crumrine D, Man MQ, Schurer N, Roelandt T, Choi EH, Uchida Y, Brown BE, Feingold KR, et al. Serine protease signaling of epidermal permeability barrier homeostasis. J Invest Dermatol. 2006; 126:2074–2086.
39. Jeong SK, Kim HJ, Youm JK, Ahn SK, Choi EH, Sohn MH, Kim KE, Hong JH, Shin DM, Lee SH. Mite and cockroach allergens activate protease-activated receptor 2 and delay epidermal permeability barrier recovery. J Invest Dermatol. 2008; 128:1930–1939.
40. Williams ML. Lipids in normal and pathological desquamation. Adv Lipid Res. 1991; 24:211–262.











 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download