1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61:69–90.
2. Cao Y, Ma J. Body mass index, prostate cancer-specific mortality, and biochemical recurrence: a systematic review and meta-analysis. Cancer Prev Res (Phila). 2011; 4:486–501.
3. Leitzmann MF, Rohrmann S. Risk factors for the onset of prostatic cancer: age, location, and behavioral correlates. Clin Epidemiol. 2012; 4:1–11.
4. Helpap B, Ringli D, Shaikhibrahim Z, Wernert N, Kristiansen G. The heterogeneous Gleason 7 carcinoma of the prostate: analyses of low and high grade (risk) carcinomas with criteria of the International Society of Urological Pathology (ISUP). Pathol Res Pract. 2013; 209:190–194.
5. Thompson I, Thrasher JB, Aus G, Burnett AL, Canby-Hagino ED, Cookson MS, D'Amico AV, Dmochowski RR, Eton DT, Forman JD, et al. AUA Prostate Cancer Clinical Guideline Update Panel. Guideline for the management of clinically localized prostate cancer: 2007 update. J Urol. 2007; 177:2106–2131.
6. Picard JC, Golshayan AR, Marshall DT, Opfermann KJ, Keane TE. The multi-disciplinary management of high-risk prostate cancer. Urol Oncol. 2012; 30:3–15.
7. Misumi T, Yamamoto Y, Murakami T, Kawai Y, Ito H, Eguchi S, Yano S, Nagao K, Hara T, Sakano S, et al. Genetic alterations at 13q14 may correlate with differences in the biological behavior of prostate cancer between Japanese and Caucasian men. Urol Int. 2010; 84:461–466.
8. Nagata D, Yoshihiro H, Nakanishi M, Naruyama H, Okada S, Ando R, Tozawa K, Kohri K. Peroxisome proliferator-activated receptor-gamma and growth inhibition by its ligands in prostate cancer. Cancer Detect Prev. 2008; 32:259–266.
9. Sikka S, Chen L, Sethi G, Kumar AP. Targeting PPARgamma signaling cascade for the prevention and treatment of prostate cancer. PPAR Res. 2012; 2012:968040.
10. Luconi M, Cantini G, Serio M. Peroxisome proliferator-activated receptor gamma (PPARγ): is the genomic activity the only answer? Steroids. 2010; 75:585–594.
11. Rosen ED, Spiegelman BM. PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001; 276:37731–37734.
12. Ota K, Ito K, Suzuki T, Saito S, Tamura M, Hayashi S, Okamura K, Sasano H, Yaegashi N. Peroxisome proliferator-activated receptor gamma and growth inhibition by its ligands in uterine endometrial carcinoma. Clin Cancer Res. 2006; 12:4200–4208.
13. Shiau CW, Yang CC, Kulp SK, Chen KF, Chen CS, Huang JW, Chen CS. Thiazolidenediones mediate apoptosis in prostate cancer cells in part through inhibition of Bcl-xL/Bcl-2 functions independently of PPARgamma. Cancer Res. 2005; 65:1561–1569.
14. Walter B, Rogenhofer S, Vogelhuber M, Berand A, Wieland WF, Andreesen R, Reichle A. Modular therapy approach in metastatic castration-refractory prostate cancer. World J Urol. 2010; 28:745–750.
15. Annicotte JS, Iankova I, Miard S, Fritz V, Sarruf D, Abella A, Berthe ML, Noël D, Pillon A, Iborra F, et al. Peroxisome proliferator-activated receptor gamma regulates E-cadherin expression and inhibits growth and invasion of prostate cancer. Mol Cell Biol. 2006; 26:7561–7574.
16. Lee NJ, Oh JH, Ban JO, Shim JH, Lee HP, Jung JK, Ahn BW, Yoon DY, Han SB, Ham YW, et al. 4-O-methylhonokiol, a PPARgamma agonist, inhibits prostate tumour growth: p21-mediated suppression of NF-kappaB activity. Br J Pharmacol. 2013; 168:1133–1145.
17. Sawayama H, Ishimoto T, Watanabe M, Yoshida N, Sugihara H, Kurashige J, Hirashima K, Iwatsuki M, Baba Y, Oki E, et al. Small molecule agonists of PPAR-gamma exert therapeutic effects in esophageal cancer. Cancer Res. 2014; 74:575–585.
18. Forootan FS, Forootan SS, Malki MI, Chen D, Li G, Lin K, Rudland PS, Foster CS, Ke Y. The expression of C-FABP and PPARgamma and their prognostic significance in prostate cancer. Int J Oncol. 2014; 44:265–275.
19. Nakamura Y, Suzuki T, Sugawara A, Arai Y, Sasano H. Peroxisome proliferator-activated receptor gamma in human prostate carcinoma. Pathol Int. 2009; 59:288–293.
20. Rogenhofer S, Ellinger J, Kahl P, Stoehr C, Hartmann A, Engehausen D, Wieland WF, Müller SC, Hofstädter F, Walter B. Enhanced expression of peroxisome proliferate-activated receptor gamma (PPAR-gamma) in advanced prostate cancer. Anticancer Res. 2012; 32:3479–3483.
21. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC cancer staging manual. 7th ed. New York: Springer;2010.
22. Jahrling JB, Hernandez CM, Denner L, Dineley KT. PPARgamma recruitment to active ERK during memory consolidation is required for Alzheimer's disease-related cognitive enhancement. J Neurosci. 2014; 34:4054–4063.
23. Ikezoe T, Miller CW, Kawano S, Heaney A, Williamson EA, Hisatake J, Green E, Hofmann W, Taguchi H, Koeffler HP. Mutational analysis of the peroxisome proliferator-activated receptor gamma gene in human malignancies. Cancer Res. 2001; 61:5307–5310.
24. Burgermeister E, Seger R. MAPK kinases as nucleo-cytoplasmic shuttles for PPARgamma. Cell Cycle. 2007; 6:1539–1548.
25. Burgermeister E, Seger R. PPARgamma and MEK Interactions in Cancer. PPAR Res. 2008; 2008:309469.
26. Demichelis F, Stanford JL. Genetic predisposition to prostate cancer: update and future perspectives. Urol Oncol. 2015; 33:75–84.
27. Jiang M, Strand DW, Franco OE, Clark PE, Hayward SW. PPARγ: a molecular link between systemic metabolic disease and benign prostate hyperplasia. Differentiation. 2011; 82:220–236.
28. Akinyeke TO, Stewart LV. Troglitazone suppresses c-Myc levels in human prostate cancer cells via a PPARgamma-independent mechanism. Cancer Biol Ther. 2011; 11:1046–1058.
29. Mueller E, Smith M, Sarraf P, Kroll T, Aiyer A, Kaufman DS, Oh W, Demetri G, Figg WD, Zhou XP, et al. Effects of ligand activation of peroxisome proliferator-activated receptor gamma in human prostate cancer. Proc Natl Acad Sci U S A. 2000; 97:10990–10995.
30. Kimura O, Kondo Y, Shimosegawa T. PPAR could contribute to the pathogenesis of hepatocellular carcinoma. PPAR Res. 2012; 2012:574180.
31. Giaginis C, Politi E, Alexandrou P, Sfiniadakis J, Kouraklis G, Theocharis S. Expression of peroxisome proliferator activated receptor-gamma (PPAR-gamma) in human non-small cell lung carcinoma: correlation with clinicopathological parameters, proliferation and apoptosis related molecules and patients' survival. Pathol Oncol Res. 2012; 18:875–883.
32. Segawa Y, Yoshimura R, Hase T, Nakatani T, Wada S, Kawahito Y, Kishimoto T, Sano H. Expression of peroxisome proliferator-activated receptor (PPAR) in human prostate cancer. Prostate. 2002; 51:108–116.
33. Lee K, Chae JY, Kwak C, Ku JH, Moon KC. TMPRSS2-ERG gene fusion and clinicopathologic characteristics of Korean prostate cancer patients. Urology. 2010; 76:1268.e7–1268.e13.
34. Miyagi Y, Sasaki T, Fujinami K, Sano J, Senga Y, Miura T, Kameda Y, Sakuma Y, Nakamura Y, Harada M, et al. ETS family-associated gene fusions in Japanese prostate cancer: analysis of 194 radical prostatectomy samples. Mod Pathol. 2010; 23:1492–1498.
35. Zmuda JM, Modugno F, Weissfeld JL, Cauley JA, Trump DL, Moffett SP, Ferrell RE. Peroxisome proliferator-activated receptor-gamma polymorphism, body mass and prostate cancer risk: evidence for gene-environment interaction. Oncology. 2006; 70:185–189.
36. Song C, Ro JY, Lee MS, Hong SJ, Chung BH, Choi HY, Lee SE, Lee E, Kim CS, Ahn H. Prostate cancer in Korean men exhibits poor differentiation and is adversely related to prognosis after radical prostatectomy. Urology. 2006; 68:820–824.
37. Goldstein NS, Hewitt SM, Taylor CR, Yaziji H, Hicks DG. Members of Ad-Hoc Committee On Immunohistochemistry Standardization. Recommendations for improved standardization of immunohistochemistry. Appl Immunohistochem Mol Morphol. 2007; 15:124–133.

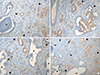
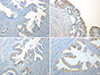


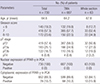
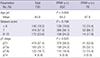




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download