1. Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001; 2:169–178.
2. Groothuis TA, Dantuma NP, Neefjes J, Salomons FA. Ubiquitin crosstalk connecting cellular processes. Cell Div. 2006; 1:21.
3. Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000; 19:94–102.
4. Park CW, Ryu KY. Cellular ubiquitin pool dynamics and homeostasis. BMB Rep. 2014; 47:475–482.
5. Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009; 10:550–563.
6. Brahimi-Horn C, Pouyssegur J. When hypoxia signalling meets the ubiquitin-proteasomal pathway, new targets for cancer therapy. Crit Rev Oncol Hematol. 2005; 53:115–123.
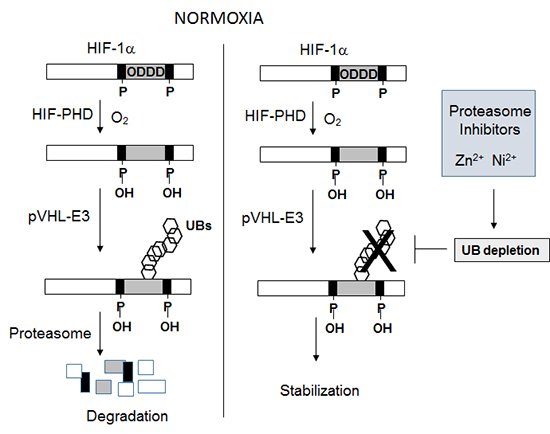
7. Chun YS, Kim MS, Park JW. Oxygen-dependent and -independent regulation of HIF-1alpha. J Korean Med Sci. 2002; 17:581–588.
8. Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004; 5:343–354.
9. Kallio PJ, Wilson WJ, O'Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem. 1999; 274:6519–6525.
10. Shin DH, Li SH, Chun YS, Huang LE, Kim MS, Park JW. CITED2 mediates the paradoxical responses of HIF-1alpha to proteasome inhibition. Oncogene. 2008; 27:1939–1944.
11. Chun YS, Choi E, Kim GT, Lee MJ, Lee MJ, Lee SE, Kim MS, Park JW. Zinc induces the accumulation of hypoxia-inducible factor (HIF)-1alpha, but inhibits the nuclear translocation of HIF-1beta, causing HIF-1 inactivation. Biochem Biophys Res Commun. 2000; 268:652–656.
12. Wang GC, Hazra TK, Mitra S, Lee HM, Englander EW. Mitochondrial DNA damage and a hypoxic response are induced by CoCI2 in rat neuronal PC12 cells. Nucleic Acids Res. 2000; 28:2135–2140.
13. Salnikow K, An WG, Melillo G, Blagosklonny MV, Costa M. Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis. 1999; 20:1819–1823.
14. Xu Q, Farah M, Webster JM, Wojcikiewicz RJ. Bortezomib rapidly suppresses ubiquitin thiolesterification to ubiquitin-conjugating enzymes and inhibits ubiquitination of histones and type I inositol 1,4,5-trisphosphate receptor. Mol Cancer Ther. 2004; 3:1263–1269.
15. Melikova MS, Kondratov KA, Kornilova ES. Two different stages of epidermal growth factor (EGF) receptor endocytosis are sensitive to free ubiquitin depletion produced by proteasome inhibitor MG132. Cell Biol Int. 2006; 30:31–43.
16. Kim I, Kim CH, Kim JH, Lee J, Choi JJ, Chen ZA, Lee MG, Chung KC, Hsu CY, Ahn YS. Pyrrolidine dithiocarbamate and zinc inhibit proteasome-dependent proteolysis. Exp Cell Res. 2004; 298:229–238.
17. Qiu M, Chen Y, Chu Y, Song S, Yang N, Gao J, Wu Z. Zinc ionophores pyrithione inhibits herpes simplex virus replication through interfering with proteasome function and NF-kappaB activation. Antiviral Res. 2013; 100:44–53.
18. Cvek B, Milacic V, Taraba J, Dou QP. Ni(II), Cu(II), and Zn(II) diethyldithiocarbamate complexes show various activities against the proteasome in breast cancer cells. J Med Chem. 2008; 51:6256–6258.
19. Frezza M, Hindo SS, Tomco D, Allard MM, Cui QC, Heeg MJ, Chen D, Dou QP, Verani CN. Comparative activities of nickel(II) and zinc(II) complexes of asymmetric [NN'O] ligands as 26S proteasome inhibitors. Inorg Chem. 2009; 48:5928–5937.







 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download