Abstract
Glutathione S-transferases (GSTs) are enzymes which play an important role in the neutralization of toxic compounds and eradication of electrophilic carcinogens. Genetic polymorphisms within the genes encoding for GSTs may therefore cause variations in their enzyme activity, which may in turn influence the interindividual susceptibility to cancers. In this study, we aimed to investigate the association between genetic polymorphisms of GSTT1, GSTM1, and GSTP1 and the risk of colorectal cancer (CRC) in 264 cases and 317 controls in a Chinese population. Genotyping was performed by using multiplex PCR (for GSTT1 and GSTM1) and PCR-RFLP (for GSTP1) methods. The association between the polymorphic genotypes and CRC risk was evaluated by deriving odds ratios (ORs) and 95% confidence intervals (CIs) using unconditional logistic regression analysis. Our results showed that individuals with GSTT1 and GSTM1 null genotypes exhibited a higher risk of CRC (GSTT1, OR,1.66; 95% CI, 1.20-2.31, P=0.003; GSTM1, OR,1.57; 95% CI,1.13-2.18, P=0.007), while no association was observed for GSTP1 (Pheterozygous=0.790 or Pvariant=0.261). Furthermore, individuals who simultaneously carried the null genotypes for both GSTT1 and GSTM1 showed a stronger risk association (OR, 1.95; 95% CI, 1.33-2.85; P<0.001). In conclusion, the GSTT1 and GSTM1 polymorphisms, but not GSTP1, may modulate the CRC risk among Chinese.
Colorectal cancer (CRC) is a leading common cancer in China and in the world (1). In many countries, the incidence of CRC shows no sign of declining and this contributes to an increasing burden of cancer-related morbidity and mortality. Although it has been well-established that exposure to environmental carcinogens is the major risk factor in the etiology of CRC, not all individuals exposed to these carcinogens develop the cancer. This indicates that CRC is multifactorial in nature, and the risk of developing CRC is influenced not only by environmental factors, but also by an underlying genetic predisposition (2).
Among the important factors affecting CRC predisposition, low penetrance genetic polymorphisms have received increasing attention in the last decade (3, 4, 5). Such polymorphisms could influence the function of the protein products encoded, and if these polymorphisms occur to the genes implicated in carcinogenesis, the interindividual variation in the functioning of the protein products could mediate the susceptibility of different individuals to the development of cancer. Considering the fact that environmental carcinogens constitute the most important risk factors for CRC, genetic polymorphisms of genes in the xenobiotic-metabolizing pathways have been widely investigated for their role in influencing CRC risk (6, 7, 8).
Glutathione S-transferases (GSTs) represent a superfamily of phase II cellular detoxification enzymes, which are involved in the metabolism of xenobiotics. Specifically, GSTs catalyze the conjugation between glutathione and electrophilic xenobiotics, thereby eliminating the harmful mutagenic or carcinogenic compounds present in diet and tobacco smoke. Several classes of GSTs have been identified, with GSTT1, M1 and P1 being the most well-characterized forms (9, 10). Polymorphisms within the genes encoding for the three GSTs, specifically GSTT1 and GSTM1 null polymorphisms as well as GSTP1 Ile105Val polymorphism, have been reported to cause reduction or total elimination of their enzymatic function (9, 11). With these in mind, we hypothesized that the mentioned GSTT1, GSTM1, and GSTP1 polymorphisms could influence the risk of CRC. Limited data is currently available on the polymorphic frequency of these genes in the Chinese population and hence, the associated CRC risk in the population. Therefore, the present study was undertaken to investigate the frequencies of GSTT1, GSTM1, and GSTP1 polymorphic genotypes among Chinese and their association with the risk of CRC.
A total of 264 cases (137 males, 127 females) and 317 controls (165 males, 152 females) were recruited into the study. The subjects were recruited between July 2012 and December 2013 from Shandong Cancer Hospital and Institute. Cases were histopathologically confirmed CRC patients, whereas controls were healthy individuals randomly selected from a cancer screening program. Controls were age- and sex-matched to the cases in terms of frequency. The demographic characteristics of the subjects are shown in Table 1.
Genomic DNA was extracted from the blood samples obtained using EasyPure Blood Genomic DNA Kit (TransGen Biotech, Beijing, China). The GSTT1 and GSTM1 null polymorphisms were genotyped by using multiplex polymerase chain reaction (PCR) technique with the beta-globin gene served as the internal control. The primers used for GSTT1 genotyping were 5'-TTCCTTACTGGTCCTCACATCTC-3' and 5'-TCACCGGATCATGGCCAGCA-3', which produced a fragment of 459 bp. On the other hand, the primers used for GSTM1 genotyping were 5'-GAACTCCCTGAAAAGCTAAAGC-3' and 5'-GTTGGGCTCAAATATACGGTGG-3', which produced a fragment of 219 bp. The 268 bp beta-globin gene was amplified by using 5'-CAACTTCATCCACGTTCACC-3' and 5'-GAAGAGCCAAGGACAGTTAC-3' primers. The multiplex PCR was performed under the following conditions: 5 min of initial denaturation at 94℃, followed by 35 cycles of 1 min at 94℃, 30 sec at 56℃, 45 sec at 72℃, and a final extension for 5 min at 72℃. The PCR products were separated on 3% agarose gels for visualization of the bands for inferring of the genotypes (Fig. 1).
On the other hand, the GSTP1 polymorphism was genotyped using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. The PCR primers used were 5'-GTAGTTTGCCCAAGGTCAAG-3' and 5'-AGCCACCTGAGGGGTAAG-3'. The PCR condition was 5 min of initial denaturation at 94℃, followed by 35 cycles of 1 min at 94℃, 30 sec at 58℃, 30 sec at 72℃, and a final extension for 5 min at 72℃. The amplification gave a product of 433 bp in size. The PCR products were then digested by using BsmAI restriction enzyme. This generated a 329 bp and a 104 bp fragment for the homozygous wild type (Ile/Ile) genotype, and a 222 bp, a 107 bp, and a 104 bp fragment for the homozygous variant (Val/Val), with the last two fragments appeared as a single band due to the small size difference (Fig. 1). Heterozygous variant genotype (Ile/Val) was detected by the presence of all the above bands in agarose gel. For all polymorphisms, approximately 10% of the samples were chosen at random and sequenced to confirm the genotypes.
Difference in genotype distribution between cases and controls was calculated by using a chi-square test. The odds ratios (ORs) and 95% confidence intervals (CI) were calculated by using unconditional logistic regression analysis to evaluate the association between the polymorphisms and CRC risk. P values of less than 0.05 were considered statistically significant.
The study was conducted according to the Declaration of Helsinki. The study protocol was reviewed and approved by the institutional review board of Shandong Academy of Medical Sciences (Ref: SAMS/SCHI/2012/28020339, English Version). Blood samples were collected from the subjects after obtaining written informed consent.
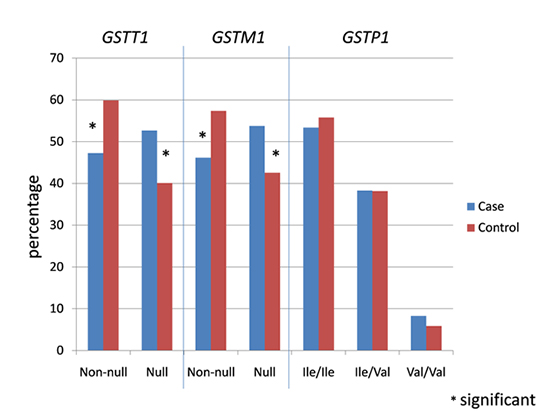
The prevalence of GSTT1, GSTM1, and GSTP1 polymorphisms is shown in Table 2. For GSTT1 and GSTM1 polymorphisms, genotype was defined as 'non-null' if at least one copy of the gene was present. Of the 264 cases, 125 (47.3%) carried non-null GSTT1 genotype and 139 (52.7%) showed null genotype. On the other hand, of the 317 controls, 190 (59.9%) and 127 (40.1%) carried non-null and null GSTT1 genotypes respectively. A significant difference was observed between cases and controls in terms of genotypic distribution (P=0.003). A significant genotypic distribution difference was also seen among cases and controls for GSTM1 polymorphism (P=0.007). A total of 122 (46.2%) of the cases and 182 (57.4%) of the controls were non-null for GSTM1. In contrast, 142 (53.8%) of the cases and 135 (42.6%) of the controls had null genotype for the polymorphism. Hardy-Weinberg test was not performed for the two polymorphisms due to the inability to distinguish between homozygous wild type and heterozygous GSTT1 and GSTM1 individuals in the analysis.
For the GSTP1 polymorphism, 141 (53.4%), 101 (38.3%) and 22 (8.3%) of the cases carried homozygous wild type (Ile/Ile), heterozygous (Ile/Val) and homozygous variant (Val/Val) genotypes respectively. A total of 177 (55.8%) of the controls were homozygous wild type for the polymorphism, while 121 (38.2%) were heterozygous, and 19 (5.9%) were homozygous variant. No significant difference in genotypic distribution was observed between cases and controls (P=0.559 for homozygous wild type, P=0.983 for heterozygous and P=0.275 for homozygous variant). The genotypic distribution did not deviate significantly from the Hardy-Weinberg equilibrium (P=0.52 for cases, P=0.78 for controls).
For GSTT1 and GSTM1 polymorphisms, the non-null genotype served as the reference for risk association analysis and was given an odds ratio (OR) value of 1. For GSTT1 polymorphisms, a significantly increased CRC risk association was observed for the null genotype (OR, 1.66; 95% CI, 1.20-2.31; P=0.003) (Table 3). Similarly, the GSTM1 null genotype also resulted in a significantly higher CRC risk association in the population studied (OR, 1.57; 95% CI, 1.13-2.18; P=0.007) (Table 3).
On the other hand, for GSTP1 polymorphism, the homozygous wild type (Ile/Ile) genotype served as the reference. The heterozygous Ile/Val genotype showed a slightly increased risk of CRC (OR, 1.05; 95% CI, 0.74-1.48) (Table 3). However, the risk was not statistically significant (P=0.790). Similarly, a non-statistically significant increased CRC risk association was observed for GSTP1 homozygous variant genotype (OR, 1.45; 95% CI, 0.76-2.80, P=0.261).
Since GSTT1 and GSTM1 polymorphisms resulted in a significantly higher CRC risk, we investigated the combined effect of the two polymorphisms in influencing the risk of CRC. In the combination analysis, individuals who simultaneously carried the non-null genotypes for both polymorphisms served as the reference. The various GSTT1 and GSTM1 combinations and their associations with CRC risk is shown in Table 4. The GSTT1 non-null and GSTM1 null combination resulted in a significantly decreased CRC risk (OR, 0.55; 95% CI, 0.32-0.97; P=0.038). Similarly, a decreased CRC risk was observed for the combination of GSTT1 null and GSTM1 non-null genotypes, although the association was not significant (OR, 0.71; 95% CI, 0.40-1.25; P=0.238). On the other hand, the GSTT1 null and GSTM1 null genotypes, which resulted in a significantly increased risk when analyzed separately, showed a stronger risk association when analyzed in combination (OR, 1.95; 95% CI, 1.33-2.85; P<0.001) (Table 4).
GSTs constitute a class of detoxification enzymes which play a pivotal role in the metabolism of carcinogens, environmental pollutants, and chemotherapeutic drugs. The enzymes have received much attention in CRC studies due to their ability to neutralize several carcinogens known to associate with the cancer, such as heterocyclic aromatic amines (HAAs) and polycyclic aromatic hydrocarbons (PAHs) (12). Several functional polymorphisms within the genes encoding for GSTT1, M1 and P1 have been described. The GSTT1 and GSTM1 null polymorphisms could lead to the total elimination of the activity of GSTT1 and M1 enzymes respectively, due to homozygosity for deletion of these genes (13). On the other hand, the GSTP1 Ile105Val substitution polymorphism could result in a decreased enzymatic activity of the protein product (14). The impaired functioning of these enzymes could present an increased risk of CRC to the individuals carrying the variant genotypes, and data on the distribution of these polymorphisms are important for the risk estimation.
The frequencies of these polymorphisms, and therefore the associated risk values, have been shown to differ from ethnicity to ethnicity and from population to population. Limited data is currently available regarding the prevalence of these polymorphisms in CRC patients and cancer-free controls in China. More often than not, studies investigating the prevalence of these polymorphisms in the above population are of small sample sizes, and did not incorporate the polymorphisms of all the three genes simultaneously (15, 16). Since GSTT1, M1, and P1 enzymes share common substrates, studies which do not include all the three genes in their analyses may result in misleading interpretations, as deficiency in one form of GST may be compensated by the presence of the other forms of GST. The present study was therefore undertaken to address the scarcity of these data.
It has been previously reported that the prevalence of GSTT1 null genotype ranged from 16%-64% in Asian populations (13). Our results were in agreement with this observation. The distribution of GSTT1 polymorphic genotypes among CRC patients in our study was highly similar to that reported by Yang et al. (6), who investigated the frequency of the polymorphism in the same population. The prevalence of non-null and null GSTT1 genotypes in our study were 47.3% and 52.7% respectively, whereas those reported by Yang et al. (6) were 49.1% and 50.9% respectively. These results were, however, not in agreement with the frequencies reported by Koh et al. (7) in a Singaporean Chinese population, which showed that the non-null genotype was more prevalent than null genotype in the population (61.3% vs. 38.8%). The genotype distribution among controls in our study was also in agreement with Yang et al. (6), in that the prevalence of non-null genotype was higher. However, the prevalence of non-null and null genotypes reported by Yang et al. (6) were 51.1% and 48.9% respectively, while ours were 59.9% and 40.1%, which were closer to the frequency reported by Koh et al. (7), i.e. 59.2% non-null genotype and 40.8% null genotype. We observed a statistically significant difference in the genotypic distribution between cases and controls in our study, while Yang et al. (6) and Koh et al. (7) reported no significant difference in the genotypic distribution. We further found a risk association between the null genotype of the polymorphism with CRC risk, an observation which was not seen in the two mentioned research groups, but was in concordance with Wang et al. (17) in India, another Asian population.
For GSTM1 polymorphism, the null genotype appeared to be more prevalent than the non-null genotype among cases in our study (53.8% vs. 46.2%), which was in agreement with Yang et al. (6) but in disagreement with Koh et al. (7). However, the frequencies reported in all research groups corresponded to previous estimation of the frequency of GSTM1 null genotype among Asians, which was in the range of 33%-63% (13). Yang et al. (6) reported a frequency of 41.3% non-null genotype and 58.7% null genotype among CRC patients. On the other hand, Koh et al. (7) reported a frequency of 51.3% non-null genotype and 48.8% null genotype among the same patients. However, among the controls, our results agreed with Koh et al. (7) and disagreed with Yang et al. (6). We reported a higher frequency of non-null genotype than null genotype among controls (57.4% vs. 42.6%), while the frequencies were 54.9% vs. 45.1% for Koh et al. (7) and 41.7% vs. 58.4% for Yang et al. (6). Similar to GSTT1 polymorphism, we found a statistically significant difference in the distribution GSTM1 polymorphic genotypes between cases and controls in our study, while the other two groups reported no statistically significant difference (6, 7). We also found a significant risk association between GSTM1 null genotype and CRC risk, which also agreed to the findings reported by Wang et al. (17). Further, we showed that the combination of GSTT1 null and GSTM1 null genotypes could present greater CRC risk to their carriers.
No data are available on the prevalence of GSTP1 polymorphism in China, although there were such data for Chinese ethnicity in Singapore and Taiwan (7, 8). In our study, no statistically significant difference exists regarding the distribution of GSTP1 polymorphic genotypes among CRC patients and controls. This observation agrees to those reported in Singapore and Taiwan, which also showed a lack of association between GSTP1 polymorphism and CRC risk (7, 8). However, our observation was in contrast to those reported by Vlaykova et al. (18) in a Bulgarian population and Hezova et al. (19) in the Central European population, which showed that the GSTP1 Ile/Val heterozygous genotype could reduce CRC risk. This difference clearly indicates the importance of ethinicity in influencing CRC risk modulation by genetic polymorphisms.
In conclusion, although small sample size constituted a limitation for our study, our results suggested that GSTT1 and GSTM1 polymorphisms could increase the risk of CRC among the Chinese population. Further, a combination of GSTT1 and GSTM1 null genotypes could present a greater CRC risk for their carriers. We also demonstrated a lack of association between GSTP1 polymorphism and CRC risk among Chinese.
Figures and Tables
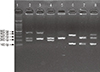 | Fig. 1Representative genotyping result. Lane 1: DNA marker. Lane 2: GSTT1 non-null, GSTM1 non-null (459 bp, 268 bp, 219 bp). Lane 3: GSTT1 non-null, GSTM1 null (459 bp, 268 bp). Lane 4: GSTT1 null, GSTM1 non-null (268 bp, 219 bp). Lane 5: GSTT1 null, GSTM1 null (268 bp). Lane 6: GSTP1 Ile/Ile (329 bp, 104 bp). Lane 7: GSTP1 Val/Val (222 bp, 107 bp, 104 bp). Lane 8: GSTP1 Ile/Val (329 bp, 222 bp, 107 bp, 104 bp). |
References
1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008 v1.2, Cancer incidence and mortality worldwide: IARC CancerBase No. 10. accessed on 1 October 2014. Available at http://globocan.iarc.fr.
2. de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004; 4:769–780.
3. Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008; 299:2423–2436.
4. Mohd Suzairi MS, Tan SC, Ahmad Aizat AA, Mohd Aminudin M, Siti Nurfatimah MS, Andee ZD, Ankathil R. The functional -94 insertion/deletion ATTG polymorphism in the promoter region of NFKB1 gene increases the risk of sporadic colorectal cancer. Cancer Epidemiol. 2013; 37:634–638.
5. Tan SC, Suzairi MS, Aizat AA, Aminudin MM, Nurfatimah MS, Bhavaraju VM, Biswal BM, Ankathil R. Gender-specific association of NFKBIA promoter polymorphisms with the risk of sporadic colorectal cancer. Med Oncol. 2013; 30:693.
6. Yang G, Gao YT, Shu XO, Cai Q, Li GL, Li HL, Ji BT, Rothman N, Dyba M, Xiang YB, et al. Isothiocyanate exposure, glutathione S-transferase polymorphisms, and colorectal cancer risk. Am J Clin Nutr. 2010; 91:704–711.
7. Koh WP, Nelson HH, Yuan JM, Van den Berg D, Jin A, Wang R, Yu MC. Glutathione S-transferase (GST) gene polymorphisms, cigarette smoking and colorectal cancer risk among Chinese in Singapore. Carcinogenesis. 2011; 32:1507–1511.
8. Yeh CC, Hsieh LL, Tang R, Chang-Chieh CR, Sung FC. Vegetable/fruit, smoking, glutathione S-transferase polymorphisms and risk for colorectal cancer in Taiwan. World J Gastroenterol. 2005; 11:1473–1480.
9. Strange RC, Fryer AA. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci Publ. 1999; 231–249.
10. Moyer AM, Salavaggione OE, Hebbring SJ, Moon I, Hildebrandt MA, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Glutathione S-transferase T1 and M1: gene sequence variation and functional genomics. Clin Cancer Res. 2007; 13:7207–7216.
11. Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997; 6:733–743.
12. Hirvonen A. Genetic factors in individual responses to environmental exposures. J Occup Environ Med. 1995; 37:37–43.
13. Cotton SC, Sharp L, Little J, Brockton N. Glutathione S-transferase polymorphisms and colorectal cancer: a HuGE review. Am J Epidemiol. 2000; 151:7–32.
14. Zimniak P, Nanduri B, Pikula S, Bandorowicz-Pikuła J, Singhal SS, Srivastava SK, Awasthi S, Awasthi YC. Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem. 1994; 224:893–899.
15. Chen K, Huang LR. A study on GSTT1 gene polymorphism and colorectal cancer susceptibility in smokers. Chin Foreign Med Res. 2009; 7:19–21.
16. Luo JG, He MJ, Liu XH. Relationship between polymorphisms in glutathione-S-transferase M1 gene and susceptibility to colorectal cancer. Anat Res. 2006; 28:52–54.
17. Wang J, Jiang J, Zhao Y, Gajalakshmi V, Kuriki K, Suzuki S, Nagaya T, Nakamura S, Akasaka S, Ishikawa H, et al. Genetic polymorphisms of glutathione S-transferase genes and susceptibility to colorectal cancer: a case-control study in an Indian population. Cancer Epidemiol. 2011; 35:66–72.
18. Vlaykova T, Miteva L, Gulubova M, Stanilova S. Ile105Val GSTP1 polymorphism and susceptibility to colorectal carcinoma in Bulgarian population. Int J Colorectal Dis. 2007; 22:1209–1215.
19. Hezova R, Bienertova-Vasku J, Sachlova M, Brezkova V, Vasku A, Svoboda M, Radová L, Kiss I, Vyzula R, Slaby O. Common polymorphisms in GSTM1, GSTT1, GSTP1, GSTA1 and susceptibility to colorectal cancer in the Central European population. Eur J Med Res. 2012; 17:17.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download