Abstract
Long QT syndrome (LQTS) is characterized by the prolongation of the QT interval in ECG and manifests predisposition to life threatening arrhythmia which often leads to sudden cardiac death. We encountered a 3-generation family with 5 affected family members in which LQTS was inherited in autosomal dominant manner. The LQTS is considered an ion channel disorder in which the type and location of the genetic mutation determines to a large extent the expression of the clinical syndrome. Upon screening of the genomic sequences of cardiac potassium ion channel genes, we found a single nucleotide C deletion mutation in the exon 3 of KCNH2 gene that co-segregates with the LQTS in this family. This mutation presumably resulted in a frameshift mutation, P151fs+15X. This study added a new genetic cause to the pool of mutations that lead to defected potassium ion channels in the heart.
Arrhythmic disorders are responsible for 2,000-3,000 deaths out of 55,000-60,000 cardiovascular disorder related death per year during last decade in Korea. One of the arrhythmic disorders, long QT syndrome (LQTS), is an inherited disorder characterized by the prolongation of the QT interval in the electrocardiogram (ECG), syncope due to ventricular tachyarrhythmia, and sudden cardiac death (1). The normal QT interval is <430 ms in male and <450 ms in female. Thus, the longer duration than these intervals (>450 ms for male and >470 ms for female) were considered disease status. Prolongation of QT interval is caused by decrease of repolarizing power during cardiac action potential, which can be resulted from either decrease in repolarizing current (late input of potassium ion) or increase in membrane current (sodium/calcium channel) during action potential plateau.
The most common causes of LQTS are mutations in the genes KCNQ1 (LQT1), HERG/KCNH2 (LQT2), and SCN5A (LQT3), which form potassium channels (KCNQ1 and KCNH2) and sodium channel (SCN5A). Mutations in the potassium channel genes, KCNQ1 and KCNH2 account for 42%-45% and 35%-45% of all LQTs with genetic mutations of congenital LQTS families, respectively (2). Mutations in SCN5A gene are identified in 2%-8% of genetic LQTS. Mutations are also found rarely in KCNE1 (LQT5), KCNE2 (LQT6), KCNJ2 (LQT7), and CACNA1c (LQT8). In addition to these genes which encodes for subunits of the cardiac ion channels thus functions to flow cardiac ion currents, Iks, Ikr, and Ina, non-channel associated genes, ankyrin B, caveolin 3, A-kinase anchor protein 9 and syntrophin, alpha 1, were also associated with LQTS (3, 4).
LQT1 (KCNQ1) gene is located in the chromosome 11p15.5 and encodes a voltage gated potassium channel KvLQT1 which coassembles with minK beta subunit (KCNE1) to form the IKs ion channel. This channel is responsible for the slow delayed rectifier potassium channel. LQT2 is caused by mutations of the potassium voltage-gated channel, subfamily H (eag-related), member 2 (KCNH2/HERG) on chromosome 7. KCNH2 codes for a protein known as Kv11.1 potassium ion channel and homotetramer of KCNH2 coassembles with Mink-related protein1 (MiRP1/KCNE2) to form IKr and conducts potassium ions out of the cardiac myocytes. Mutations of the alpha subunit of sodium channel gene, SCN5A, cause LQT3. This gene is located at chromosome 3p21-24.
LQTS is inherited mainly in dominant fashion with rare cases of recessive inheritance seen in Jervell Lange-Nielsen syndrome (5) Much progress in congenital long QT syndrome has been made with availability of the molecular diagnostic tools and better understanding of the relationship between genotype and phenotype. In this study we analyzed the KCNQ1, KCNH2, and KCNE1 gene of a Korean family with LQT syndrome and discovered a genetic mutation co-segregating with the disorder.
The proband who was admitted at the St. Mary's Hospital on May 29th, 2001 was a 39-yr-old female patient referred for the evaluation of intermittent palpitation and dizziness. Clinical evaluation including QT interval assessment and T-wave morphology through ECG, 24 hr Holter monitoring, treadmill exercise test, and coronary angiography was performed. Pedigree analysis was also carried out. The QT intervals were measured by Ashman method and corrected for heart rate (QTc) (6). All the family members were also evaluated by ECG.
The patient complained severe dizziness and syncope with palpitation when she was under the stress or emotionally upset. Twenty-four hour Holter monitoring showed a few premature atrial or ventricular complexes. During treadmill exercise test, horizontal ST depressions were appeared on leads II, III, avF, V4-V5 with chest pain. While we are taking coronary angiography, the patient complained palpitation, dizziness, and loss of consciousness for a few seconds. Then the ECG showed typical 'Torsades de Pointes' with T wave alternans (Fig. 1). She was diagnosed as long QT syndrome and prescribed propranolol 40 mg bid. After medication, the symptom improved and she was discharged with good condition.
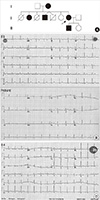
Fig. 2A shows the pedigree of this LQTS family. Five family members (I2, II2, II4, II7, and III1) were diagnosed with LQTS using the diagnostic scoring system (7). The rest 4 members showed normal ECG patterns (I1, II6, II8, and III2) with normal QTc intervals (<450 ms) and the representative ECG is shown (Fig. 2B). With exception of subject II4, all affected members showed prolonged QTc intervals ranged from 454 to 527 ms and the proband's ECG is shown in Fig. 2B. Subject II4 was diagnosed as LQTS based on the non-specific T-wave abnormality despite of shorter QTc interval than normal one (Fig. 2C). The clinical characteristics are summarized in Table 1.
Genomic DNA was extracted from the peripheral blood of the participants using a Blood extraction kit (Qiagen) following the manufacturer's protocol. Genomic DNA was used as a template for PCR amplification of the individual exons of the KCNQ1, KCNH2, and KCNE1 genes. Gene-specific primer sequences were designed using genomic sequences of each gene (Genbank accession numbers; NG_008935.1 for KCNQ1, NG_008916.1 for KCNH2, and NG_009091.1for KCNE1). PCR primer sequences and conditions are shown in the Table 2. The purified PCR product was directly sequenced using ABI Prism 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA) and the sequences analyzed using BLAST program (http://blast.ncbi.nlm.nih.gov/). The putative mutation was further investigated in the family members to determine its familial transmission. And the general Korean population was also screened for the presence of this mutation.
The sequencing of the entire coding regions as well as the splicing junctions of the KCNQ1, KCNH2, and KCNE1 genes of the proband revealed a typical chromatogram of mixed samples in the sequence read of an exon (Fig. 3A). Subcloning of the PCR products followed by sequencing of individual clones identified a mutant allele with a single nucleotide C deletion in exon 3 of KCHN2 gene. This C deletion at the nucleotide position of 453 of the KCNH2 mRNA presumably resulted in frameshift of the coding region thus introducing a premature termination codon after 15 miscoded amino acids (151Pfs+15X) (Fig. 3B). This mutation co-segregated with LQTS in this family, thus I2, II2, II4, II7 (proband), and III1 were all heterozygotes for delC453fs (151Pfs+15X). This mutation was not detected in 100 chromosomes in general Korean population.
The clinical course of patients with LQTS is so variable and is affected by the length of QT interval, sex and the genotype (8). The risk of sudden cardiac death is significantly higher among patients with LQT1 and LQT2 mutations than those with LQT3 mutation (9). Sudden cardiac death usually occurs under specific circumstances in a gene-specific manner. Patients with LQT2 usually experience cardiac events with acute arousal-type emotions, such as the ringing of an alarm clock (10, 11).
A voltage-gated K+ channel which flaws the rapidly activating delayed rectifying Ikr current is formed by tetramer of human KCNH2α subunit and MiRP1β subunit and acts to keep the balance of membrane currents flow during the action potential plateau and repolarization in the heart.
The mutations in the KCNH2 gene cause LQTS2 which is the second most common cause of the congenital LQTS and responsible for 35%-45% of all genotyped LQTS. The KCNH2 gene codes for an 1159 amino acid polypeptide which folds into 6 transmembrane α helices, numbered S1-S6, a pore helix situated between S5 and S6, and cytoplasmic N- and C-termini (12).
Currently, near 300 mutations were reported in the KCNH2 gene (http://www.fsm.it/cardmoc/) for LQT2 and these mutations can lead to LQTS by different mechanisms. These mutations exert influence on Ikr by reduced synthesis of HERG, defected trafficking of KCNH2 from ER to plasma membrane, defective gating or defect in ion permeation (13, 14). Those mutations include missense mutation (64%), frameshift mutation due to deletion or insertion (~21%), nonsense mutation (8%), splice error mutation (3.5%), and inframe deletion/insertion mutation (3.5%).
In this study, we described a deletion mutation del453C in N-terminal region of the KCNH2 gene in a family with long QT syndrome. This mutation has been previously reported in the Finish LQT families (15, 16) and the affected patients exhibited the mean QTc of 470 + 42 ms. Similarly to this characteristics, all five carriers in this Korean LQTs pedigree manifested LQT phenotype with the prolonged QTc intervals with average 484.33±37.31 ms. and some with abnormal T wave phenotype while non-carriers showed normal ECG. Thus, the penetrance of LQTS in this family was assumed to be 100%. This del453C mutation presumably leads to deficiency in the rapid delayed rectifier potassium channel and resulted in the prolonged the open state of the channel thus reducing in the repolarizing current.
The mutant KCNH2 (151Pfs+15X) we found in this Korean LQTS family presumably resulted in a frameshift of the coding sequence and thus introduced a stop codon unexpectedly. Therefore, the mutant mRNA may have suffered nonsense-mediated decay of mRNA, which resulted in the decreased production of KCNH2 protein. In this case, the cells may have experienced haplo-insufficiency of protein causing a disease state. Alternate mechanism could be that the mutant allele mRNA may not be affected in its stability but it was translated and resulted in the truncated KCNH2 protein due to frame shift. In turn, this truncated protein may have caused abnormal formation of potassium channel leading to dominant negative effect.
N-terminal region of truncated KCNH2 protein (1-151 AA) contains a Per-Arnt-Sem (PAS) domain (1-137 AA) which is found in many signaling proteins and functions as a signal sensor. Functionally, PAS domain of KCNH2 was shown to modulate deactivation process of the channel (12, 17) and N-terminal 16 AA have been implicated in slowing the deactivation process and stabilizing the open state (18).
Interestingly, N-terminus (1-135AA) was shown to form a tetramer in the absence of the rest of KCNH2 protein. In addition, when it was co-expressed with wild type KCNH2 in which was the case in our patients, truncated mutant inhibited the functional expression of KCNH2 channel in transfected cells (19). Furthermore, this soluble peptide of N-terminus (1-135 AA) was shown to bind to membrane bound KCNH2 (12) suggesting that the truncated mutant KCNH2 protein may affect formation of KCNH2 tetramer leading to abnormal function of potassium channel. Currently, it is not known whether this mutation 151Pfs+15X caused expression of truncated protein and inhibited formation of functional potassium channel. Molecular approach as well as electrophysiological study with mutant form of this gene can be utilized to elucidate the mechanism underlying LQTS with this mutation.
Figures and Tables
Fig. 1
It showed typical figures of 'Torsades de Pointes', In (A), the black arrows indicate
long QT interval and T wave alternans.
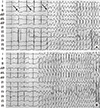
Fig. 2
Pedigree and Electrocardiogram. (A) Pedigree of the LQTS family; filled symbols represent affected members whereas empty symbols normal members. Arrow indicates the proband. (B) Electrocardiogram of normal (II6) and the proband (II7). (C) Electrocardiogram of II4 displays atypically short QTc interval than those of the other family members. This patient also contains del453 C mutation (151Pfs+15X) in the KCNH2 gene.
References
1. Moss AJ, Kass RS. Long QT syndrome: from channels to cardiac arrhythmias. J Clin Invest. 2005; 115:2018–2024.
2. Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent GM, et al. Spectrum of mutations in long-QT syndrome genes: KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000; 102:1178–1185.
3. Hedley PL, Jørgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M. The genetic basis of long QT and short QT syndromes: a mutation update. Hum Mutat. 2009; 30:1486–1511.
4. Itoh H, Shimizu W, Hayashi K, Yamagata K, Sakaguchi T, Ohno S, Makiyama T, Akao M, Ai T, Noda T, et al. Long QT syndrome with compound mutations is associated with a more severe phenotype: a Japanese multicenter study. Heart Rhythm. 2010; 7:1411–1418.
5. Jervell A, Lange-Nielsen F. Congenital deaf-mutism, functional heart disease with prolongation of the Q-T interval and sudden death. Am Heart J. 1957; 54:59–68.
6. Sagie A, Larson MG, Goldberg RJ, Bengtson JR, Levy D. An improved method for adjusting the QT interval for heart rate (the Framingham Heart Study). Am J Cardiol. 1992; 70:797–801.
7. Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome: an update. Circulation. 1993; 88:782–784.
8. Moss AJ, Schwartz PJ, Crampton RS, Tzivoni D, Locati EH, MacCluer J, Hall WJ, Weitkamp L, Vincent GM, Garson A Jr, et al. The long QT syndrome: prospective longitudinal study of 328 families. Circulation. 1991; 84:1136–1144.
9. Zareba W, Moss AJ, Schwartz PJ, Vincent GM, Robinson JL, Priori SG, Benhorin J, Locati EH, Towbin JA, Keating MT, et al. Influence of genotype on the clinical course of the long-QT syndrome: International Long-QT Syndrome Registry Research Group. N Engl J Med. 1998; 339:960–965.
10. Moss AJ, Robinson JL, Gessman L, Gillespie R, Zareba W, Schwartz PJ, Vincent GM, Benhorin J, Heilbron EL, Towbin JA, et al. Comparison of clinical and genetic variables of cardiac events associated with loud noise versus swimming among subjects with the long QT syndrome. Am J Cardiol. 1999; 84:876–879.
11. Ali RH, Zareba W, Moss AJ, Schwartz PJ, Benhorin J, Vincent GM, Locati EH, Priori S, Napolitano C, Towbin JA, et al. Clinical and genetic variables associated with acute arousal and nonarousal-related cardiac events among subjects with long QT syndrome. Am J Cardiol. 2000; 85:457–461.
12. Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998; 95:649–655.
13. Clancy CE, Rudy Y. Cellular consequences of HERG mutations in the long QT syndrome: precursors to sudden cardiac death. Cardiovasc Res. 2001; 50:301–313.
14. Thomas D, Kiehn J, Katus HA, Karle CA. Defective protein trafficking in hERG-associated hereditary long QT syndrome (LQT2): molecular mechanisms and restoration of intracellular protein processing. Cardiovasc Res. 2003; 60:235–241.
15. Laitinen P, Fodstad H, Piippo K, Swan H, Toivonen L, Viitasalo M, Kaprio J, Kontula K. Survey of the coding region of the HERG gene in long QT syndrome reveals six novel mutations and an amino acid polymorphism with possible phenotypic effects. Hum Mutat. 2000; 15:580–581.
16. Fodstad H, Swan H, Laitinen P, Piippo K, Paavonen K, Viitasalo M, Toivonen L, Kontula K. Four potassium channel mutations account for 73% of the genetic spectrum underlying long-QT syndrome (LQTS) and provide evidence for a strong founder effect in Finland. Ann Med. 2004; 36:53–63.
17. Chen J, Zou A, Splawski I, Keating MT, Sanguinetti MC. Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J Biol Chem. 1999; 274:10113–10118.
18. Wang J, Myers CD, Robertson GA. Dynamic control of deactivation gating by a soluble amino-terminal domain in HERG K(+) channels. J Gen Physiol. 2000; 115:749–758.
19. Li X, Xu J, Li M. The human delta1261 mutation of the HERG potassium channel results in a truncated protein that contains a subunit interaction domain and decreases the channel expression. J Biol Chem. 1997; 272:705–708.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download