Abstract
The innate immune response in patients who develop inflammatory bowel disease (IBD) may be abnormal. However, the exact role of Toll-like receptors (TLRs) / CD14 gene in the pathogenesis of IBD has not been fully elucidated. We aimed to investigate the association between polymorphisms of TLR1, 2, 4, 6, and CD14 gene and susceptibility to IBD in Korean population. A total 144 patients of IBD (99 patients with ulcerative colitis, 45 patients with Crohn's disease) and 178 healthy controls were enrolled. Using a PCR-RFLP, we evaluated mutations of TLR1 (Arg80Thr), TLR2 (Arg753Gln and Arg677Trp), TLR4 (Asp299Gly and Thr399Ile), TLR6 (Ser249Pro) genes and the -159 C/T promoter polymorphism of CD14 gene. No TLR polymorphisms were detected in Korean subjects. T allele and TT genotype frequencies of CD14 gene were significantly higher in IBD patients than in healthy controls. In subgroup analysis, T allelic frequency was higher in pancolitis phenotype of ulcerative colitis. In Korean population, the promoter polymorphism at -159 C/T of the CD14 gene is positively associated with IBD, both ulcerative colitis and Crohn's disease.
Inflammatory bowel disease (IBD) is a complex disease with both genetic and environmental factors necessary for disease pathogenesis. Particular genetic changes may influence the susceptibility to disease and individual variation of disease pattern. It has been associated with specific variant alleles (polymorphism) of gene that are present in a significant proportion of the normal population. Polymorphisms in a wide variety of genes may modify the effect of environmental exposures (1). Recent genome-wide linkage studies and case-control association studies have shown several susceptibility regions for IBD (2-4).
The innate immune system represents the first defense line in preventing infection of bacteria and provides a critical interface between microorganisms and their hosts. It can promptly recognize and respond to pathogenic microorganisms. Toll-like receptor (TLR) is one of pathogen pattern recognition receptors for micro-organism-derived pathogenic molecules (5, 6). Clusters of differentiation 14 (CD14) is a glycoprotein expressed on the surfaces of monocytes and macrophages, and acts as a pattern recognition receptor and contributes to TLR-induced cell activation (5, 7). Both of TLRs and CD14 are considered as the primary sensors of the innate immune system.
It has been reported that the -159 C/T polymorphism in the CD14 gene plays a significant role in regulating the CD14 expression by increasing production of TNF-α (8). Previously, several studies have reported on the association between IBD and TLRs/ CD14 gene polymorphism (9-15), but there are no consistent results. Different results may be interpreted by genetic heterogeneity of races. Until now, in Korean population, whether TLRs/CD14 gene polymorphism is associated with IBD remains unknown.
In this study, we aimed to investigate the frequencies of TLR1, 2, 4, 6, and CD14 gene polymorphism in patients with IBD, and evaluate the association between those polymorphisms and the susceptibility to IBD in Korean population.
Healthy control subjects and IBD (ulcerative colitis [UC] and Crohn's disease [CD]) patients were enrolled from a teaching hospital of the Catholic University of Medicine, St. Vincent's hospital, from September 2007 to February 2008. A diagnosis of UC was made if all the following three criteria were present: a typical history of diarrhea or blood and pus in stool for longer than 4 weeks; a typical sigmoidoscopic or colonoscopic picture with diffusely granular, friable, or ulcerated mucosa without rectal sparing; and characteristic histopathological signs of infiammation on biopsy (16). Diagnosis of CD was based on the official criteria established by the research committee of the Japan Ministry of Health and in Western countries (17, 18). Healthy control subjects were asymptomatic and randomly selected from subjects attending regular health screening at the Health Promotion Center of the same hospital. They were not affected with colon by endoscopic examination or double contrast barium enema. Blood samples of 10 mL were obtained from all subjects and collected in a test tube containing EDTA or heparin.
Chronic UC patients were classified according to the location and extension of inflammatory lesions judged by endoscopic findings, the disease activity index following Mayo score, and the need for or the response of steroid and immunomodulator therapy. Chronic CD patients were classified according to the location of lesions and the disease behavior following Montreal classification (19), and the need for or the response of steroid and immunomodulator therapy. Patients who failed to respond to steroid over a period of 4 weeks were defined as 'steroid refractory'. Patients who initially responded to steroid but then relapsed during tapering, and required re-introduction of steroid therapy to maintain symptoms control, were defined as 'steroid dependent' (20).
Genomic DNA was obtained from buffy-coat leukocytes using the AccuPrep Genomic DNA Extraction Kit (Bioneer Corporation, Daejeon, Korea). The CD14 promoter C(-159)T, TLR1 Arg80Thr, TLR2 Arg677Trp, TLR2 Arg753Gln, TLR4 Asp299Gly, TLR4 Thr399Ile, TLR6 Ser249Pro polymorphisms were performed using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method.
PCR reactions were set up using the i-Star Taq DNA polymerase (iNtRON Biotechnology, Seoul, Korea). In a total reaction volume of 20 µL, 2 µL 10 × buffer, 10 pM each primer, 2 µL genomic DNA, 2 µL dNTP mixture were combined. Reactions were run on a Bio-Rad MyCycler thermal cyclers (BIO-RAD, Philadelphia, PA, USA). The primer sequences for amplifying each SNP were shown in Table 1. The PCR conditions were as follows: an initial denaturation at 95℃ for 5 min, followed by 35 cycles of denaturing at 94℃ for 30 sec, annealing for 30 sec, extension at 72℃ for 30 sec and final incubation at 72℃ for 10 min and cooling to 4℃. For 16 hr, the PCR products (3 µL) were used for an overnight digest with the appropriate restriction enzyme (Table 1), and digests were analyzed by electrophoresis in a 3% agarose gel.
The data were analyzed using SPSS software (version 15.0, SPSS, Chicago, IL, USA). Genotype and allele frequencies were calculated by direct counting. Significance of association was determined by chi-square tests or Fisher's exact test. Departures from Hardy-Weinberg equilibrium in patients and healthy controls were tested by the chi-square method. Continuous data were expressed as mean ± standard deviation and were analyzed using independent samples t-tests. A P value < 0.05 was considered significant for all the tests. For subgroup analysis, Bonferroni correction was made for the multiple comparisons.
A total of 144 IBD patients and 178 healthy control subjects were enrolled in the study. There were no significant differences in age and sex between the two groups. The demographic characteristics of the patients with UC and CD were shown in Table 2.
Arg80Thr of TLR 1, Arg677Trp of TLR2, Arg753Gln of TLR2, Asp299Gly of TLR4, Thr399Ile of TLR4 and Ser249Pro TLR6 gene polymorphisms were not detected in healthy control subjects and IBD patients.
Digestion of the PCR products for the promoter polymorphism at -159 C/T of the CD14 gene yielded bands of 295 bp in TT homozygotes, 140 and 155 bp in CC homozygotes, and all 3 bands (140, 155, and 295 bp) in heterozygotes (Fig. 1). The frequencies of TT genotype and T allele of the CD14 gene were significantly higher in IBD patients than in healthy control group (TT, 32.6% in IBD vs 14.6% in control, P < 0.01; T allele, 0.57 in IBD vs 0.34 in healthy control, P < 0.01). The frequencies of TT and T allele in each of UC and CD patients were also significantly higher than in controls (Table 3).
A total 45 patients with CD assigned to divide into the sub-groups according to the Montreal classification. There were no significant differences in TT genotype and T allele frequencies of CD14 gene according to the disease location and behavior, the administration of steroid or immunomodulator, and the response to treatment (Table 4). Among 99 patients with UC, 24 patients (22.2%) had pancolitis, while 23 patients had the disease limited to the left side and 52 patients had proctitis. In patients with UC, TT and T allele were not associated with the disease activity, the use of steroid or immunomodulator, and the response to each drug, but associated with the disease extent (T allele: 0.65 in pancolitis vs 0.51 in left-sided colitis and proctitis, P = 0.01) (Table 5) (Fig. 2).
Although the exact etiology of IBD is unknown, it is considered that inappropriate and exaggerated immune response to luminal bacteria would be an important role in the pathogenesis of IBD. In the past, numerous endogenous, pathogenic bacteria or mycobacteria have long been suspected as being major contributors to the etiology of IBD, but none of them have achieved etiologic status (21). Inappropriate activation of innate immunity is the other arm involved in the pathogenesis of IBD. Recently, genetic studies in humans and mice have made a convincing link between the innate immune system and chronic inflammation (22, 23).
Epidemiological studies show an association between TLR4 polymorphism and susceptibility to IBD. In a Belgian study, the allele frequency of the TLR4 A299G polymorphism was significantly higher in CD and in UC patients compared with controls, whereas no association with IBD of the TLR4 polymorphim was found in Hungarian and German cohort (24, 25). The study about polymorphisms of TLR1, 2 or 6 in the pathogenesis of IBD are scarce. None of the single nucleotide polymorphisms of TLR1, 2 or 6 were involved with IBD susceptibility. However, a number of variants were found to be associated with disease phenotypes. The TLR2 R753G and TLR1 R80T single nucleotide polymorphisms (SNPs) were found to be associated with pancolitis in UC (26). In our results, gene polymorphisms of the TLRs were not detected in any Korean subject. It suggested that the contribution of genetic determinants might differ significantly between ethnicities.
CD14 gene consists of a single base substitution (C → T) at position -159 in the promoter of the gene (27). This polymorphism has been reported to influence degree of the density of CD14 expression to secrete inflammatory cytokines by lipopolysaccharide (LPS) (28). Previously, in several reports from Western countries, gene polymorphisms of the CD14 was found to be associated with the development of IBD, more so in the development of CD than in UC (9, 10). However, in Japanese population, the same genotype is associated with UC but not with CD (12, 15), and another study investigating Chinese patients failed to demonstrate any association (13). In our results, T allele and TT genotype frequencies of CD14 gene were significantly higher in IBD patients than in healthy controls. In subgroup analysis, both CD and UC patients had significantly higher T allele frequency compared to healthy controls. In patients with UC and CD, T allele and TT genotype frequencies of CD14 gene were not associated with the disease activity, the use of steroid or immunomodulator and the response to each drug. It is noteworthy that the specific allelic frequency of CD14 polymorphism is closely associated with pancolitis phenotype in UC. Since this study includes small numbers of the patients with IBD, especially with CD, it might be difficult to figure out the association between genetic polymorphism and disease phenotype. Moreover, it cannot exclude a type 2 statistical error. To overcome it, the researcher can sample as many subjects as cost and time allow.
The distributions of CD14 -159C/T genotypes were significantly different between Korean and Chinese, and between Korean and Japanese population, which demonstrated ethnic genetic differences. Compared with the previous studies, the frequency of the CC genotype of healthy controls in Korean population (41.6%) was higher than in Chinese (15.6%) or Japanese (21.8%), whereas the TT genotype of healthy controls in Korean (14.6%) was lower than in Chinese (36.3%) or Japanese (25.4%) (12, 13). Further study in larger and diverse populations would be needed for elucidation of the association between disease susceptibility of IBD and polymorphism of CD14 gene.
In conclusion, the promoter polymorphism at -159 C/T of the CD14 gene is positively associated with IBD, both UC and CD. In patients with UC, T allele and TT genotype frequencies of CD14 gene are associated with the disease extent, and increased T allelic frequency is preferable to pancolitis phenotype of UC in Korean population.
Figures and Tables
Fig. 1
Detection for the -159C/T polymorphism of CD14 gene by using PCR-RFLP. Digestion of the PCR products yielded bands of 295 bp in TT homozygotes, 140 and 155 bp in CC homozygotes, and all 3 bands (140, 155, and 295 bp) in heterozygotes.
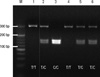
Fig. 2
Allele frequency of the promoter polymorphism at -159 C/T of the CD14 gene according to disease extent in patients with ulcerative colitis.

Table 1
The primer sequences, annealing time, restriction enzymes, and digest time used for PCR-RFLP methods detecting the each single nucleotide polymorphism (SNP)

References
1. Perera FP, Weinstein IB. Molecular epidemiology: recent advances and future directions. Carcinogenesis. 2000. 21:517–524.
2. Freeman HJ. Familial Crohn's disease in single or multiple first-degree relatives. J Clin Gastroenterol. 2002. 35:9–13.
3. Ahmad T, Satsangi J, McGovern D, Bunce M, Jewell DP. The genetics of inflammatory bowel disease. Aliment Pharmacol Ther. 2001. 15:731–748.
4. Hugot JP, Thomas G. Genome-wide scanning in inflammatory bowel disease. Dig Dis. 1998. 16:364–369.
5. Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000. 406:782–787.
6. Akira S, Sato S. Toll-like receptors and their signaling mechanisms. Scand J Infect Dis. 2003. 35:555–562.
7. Antal-Szalmás P. Evaluation of CD14 in host defence. Eur J Clin Invest. 2000. 30:167–179.
8. Lin J, Yao YM, Yu Y, Chai JK, Huang ZH, Dong N, Sheng ZY. Effects of CD14-159 C/T polymorphism on CD14 expression and the balance between proinflammatory and anti-inflammatory cytokines in whole blood culture. Shock. 2007. 28:148–153.
9. Klein W, Tromm A, Griga T, Fricke H, Folwaczny C, Hocke M, Eitner K, Marx M, Duerig N, Epplen JT. A polymorphism in the CD14 gene is associated with Crohn disease. Scand J Gastroenterol. 2002. 37:189–191.
10. Gazouli M, Mantzaris G, Kotsinas A, Zacharatos P, Papalambros E, Archimandritis A, Ikonomopoulos J, Gorgoulis VG. Association between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J Gastroenterol. 2005. 11:681–685.
11. Leung E, Hong J, Fraser AG, Merriman TR, Vishnu P, Abbott WG, Krissansen GW. Polymorphisms of CARD15/NOD2 and CD14 genes in New Zealand Crohn's disease patients. Immunol Cell Biol. 2005. 83:498–503.
12. Obana N, Takahashi S, Kinouchi Y, Negoro K, Takagi S, Hiwatashi N, Shimosegawa T. Ulcerative colitis is associated with a promoter polymorphism of lipopolysaccharide receptor gene, CD14. Scand J Gastroenterol. 2002. 37:699–704.
13. Guo QS, Xia B, Jiang Y, Morré SA, Cheng L, Li J, Crusius JB, Peña AS. Polymorphisms of CD14 gene and TLR4 gene are not associated with ulcerative colitis in Chinese patients. Postgrad Med J. 2005. 81:526–529.
14. Peters KE, O'Callaghan NJ, Cavanaugh JA. Lack of association of the CD14 promoter polymorphism: 159C/T with Caucasian inflammatory bowel disease. Scand J Gastroenterol. 2005. 40:194–197.
15. Wang F, Tahara T, Arisawa T, Shibata T, Nakamura M, Fujita H, Iwata M, Kamiya Y, Nagasaka M, Takahama K, Watanabe M, Hirata I, Nakano H. Genetic polymorphisms of CD14 and Toll-like receptor-2 (TLR2) in patients with ulcerative colitis. J Gastroenterol Hepatol. 2007. 22:925–929.
16. Yang SK, Hong WS, Min YI, Kim HY, Yoo JY, Rhee PL, Rhee JC, Chang DK, Song IS, Jung SA, Park EB, Yoo HM, Lee DK, Kim YK. Incidence and prevalence of ulcerative colitis in the Songpa-Kangdong district, Seoul, Korea, 1986-1997. J Gastroenterol Hepatol. 2000. 15:1037–1042.
17. Gasche C, Scholmerich J, Brynskov J, D'Haens G, Hanauer SB, Irvine EJ, Jewell DP, Rachmilewitz D, Sachar DB, Sandborn WJ, Sutherland LR. A simple classification of Crohn's disease: report of the Working Party for the World Congresses of Gastroenterology, Vienna 1998. Inflamm Bowel Dis. 2000. 6:8–15.
18. Caprilli R, Gassull MA, Escher JC, Moser G, Munkholm P, Forbes A, Hommes DW, Lochs H, Angelucci E, Cocco A, Vucelic B, Hildebrand H, Kolacek S, Riis L, Lukas M, de Franchis R, Hamilton M, Jantschek G, Michetti P, O'Morain C, Anwar MM, Freitas JL, Mouzas IA, Baert F, Mitchell R, Hawkey CJ. European Crohn's and Colitis Organisation. European evidence based consensus on the diagnosis and management of Crohn's disease: special situations. Gut. 2006. 55:i36–i58.
19. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006. 55:749–753.
20. Stange EF, Travis SP, Vermeire S, Beglinger C, Kupcinkas L, Geboes K, Barakauskiene A, Villanacci V, Von Herbay A, Warren BF, Garche C, Tilg H, Schreiber SW, Schölmerich J, Reinisch W. European Crohn's and Colitis Organisation. European evidence based consensus on the diagnosis and management of Crohn's disease: definition and diagnosis. Gut. 2006. 55:i1–i15.
21. Ohkusa T, Nomura T, Sato N. The role of bacterial infection in the pathogenesis of inflammatory bowel disease. Intern Med. 2004. 43:534–539.
22. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004. 118:229–241.
23. Lee J, Mo JH, Katakura K, Alkalay I, Rucker AN, Liu YT, Lee HK, Shen C, Cojocaru G, Shenouda S, Kagnoff M, Eckmann L, Ben-Neriah Y, Raz E. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. 2006. 8:1327–1336.
24. Baumgart DC, Buning C, Geerdts L, Schmidt HH, Genschel J, Fiedler T, Gentz E, Molnar T, Nagy F, Lonovics J, Lochs H, Wiedenmann B, Nickel R, Witt H, Dignass A. The c.1-260C>T promoter variant of CD14 but not the c.896A>G (p.D299G) variant of toll-like receptor 4 (TLR4) genes is associated with inflammatory bowel disease. Digestion. 2007. 76:196–202.
25. Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, Quertinmont E, Abramowicz M, Van Gossum A, Devière J, Rutgeerts P. Deficient host-bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut. 2004. 53:987–992.
26. Pierik M, Joossens S, Van Steen K, Van Schuerbeek N, Vlietinck R, Rutgeerts P, Vermeire S. Toll-like receptor-1,-2,and-6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm Bowel Dis. 2006. 12:1–8.
27. Hubacek JA, Rothe G, Pit'ha J, Skodová Z, Stanëk V, Poledne R, Schmitz G. C(-260)→T polymorphism in the promoter of the CD14 monocyte receptor gene as a risk factor for myocardial infarction. Circulation. 1999. 99:3218–3220.
28. Schütt C. Fighting infection: the role of lipopolysaccharide binding proteins CD14 and LBP. Pathobiology. 1999. 67:227–229.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download