See the reply "The Author Response: EML4-ALK Fusion Gene in Korean Non-Small Cell Lung Cancer" in Volume 27 on page 578.
We read with interest the article "EML4-ALK Fusion Gene in Korean Non-Small Cell Lung Cancer" in a recent issue of the Journal of Korean Medical Science by Jin et al. (1). In this study, EML4-ALK fusion gene was detected in 10 of 167 non-small cell lung cancer (NSCLC) patients by using reverse transcriptase-polymerase chain reaction (RT-PCR) as a basic screening technique instead of fluorescence in situ hybridization (FISH) method. The frequency of EML4-ALK fusion gene in this study (6.0%) was not largely different from a previous study that used the FISH method on Korean NSCLC patients (4.2%) (2), while it did not diverge from the results in previous studies on a general NSCLC patient population that yielded 3%-7% ratios as well. Based on the experience of detecting leukemia fusion genes with several new molecular methods during the diagnosis of leukemia (3-6), we would like to mention the pros and cons of such methods in the analysis of EML4-ALK fusion genes and introduce the usefulness of long distance- (LD-) or long distance inverse-polymerase chain reaction (LDI-PCR) on solid tumors including lung cancer.
As correctly mentioned by Jin et al. (1), the shortcomings of ALK breakapart FISH test include its inability to identify accurate subtypes of the EML4-ALK fusion gene and discriminate the changes in ALK partner genes, which is due to the characteristics of the breakapart FISH probe. To confirm the presence of a partner gene or the fusion gene subtype, additional methods such as PCR would be always necessary. In that sense, the fact that Jin et al. (1) used RT-PCR as a molecular screening method was a good approach to overcome the above mentioned limitations of FISH. However, it should be noted that previous studies have indicated a number of variant EML4-ALK fusion gene subtypes which could be missed if not designing a complete set of multiplex oligonucleotides (7). This is one of the important limitations of RT-PCR in fusion gene analysis that Jin et al. (1) have overlooked. Furthermore, ALK gene has additional fusion partners such as TGF, KIF5B, and KLC1 in lung cancer (8).
We believe that the established methodologies concerning MLL rearrangements in leukemia patients could be a prototypic example of similar technical pitfalls. As widely known, MLL rearrangements have more than 70 partner genes while each MLL rearrangement can also show various types of fusion genes due to the involvement of different introns of the MLL- and the corresponding partner gene (3). As mentioned in our recent paper, "Diagnostic standardization of leukemia fusion gene detection system using multiplex reverse transcriptase-polymerase chain reaction in Korea," (6) these characteristics limit a complete detection of all MLL rearrangements in any multiplex RT-PCR method that has been developed so far. This is also pertinent to the reason why FISH has been used as a screening method in various leukemia fusion gene analyses, despite its aforementioned limitations. Moreover, another important limitation of the RT-PCR method is that it uses RNA and a subsequent complementary DNA (cDNA) as clinical specimens that are relatively unstable (compared to genomic DNA). In fusion gene analyses on hematologic malignancies, it is not uncommon to encounter such a situation where an alternative method is required because of the unstable nature of RNA samples as well as the unavailability of the sample in the first place. Recently, we also have been trying to resolve such limitations of FISH and RT-PCR methods in our molecular analyses on leukemia patients that showed fusion genes other than MLL rearrangements by using a LD-/LDI-PCR method (9-11).
The principles of LD-/LDI-PCR were explained in our previous papers in detail (3, 4). To briefly elaborate, LD-/LDI-PCR is a fusion breakpoint analysis method that uses stable genomic DNA and incorporates both the sensitivity of the FISH method that can screen all types of fusion genes and the specificity of the RT-PCR method that can confirm specific subtypes of a fusion gene. Fusion gene analysis using such LD-/LDI-PCR method has not only been successfully implemented in the diagnostic schemes for leukemia patients but has turned out to be informative also on solid tumors, like e.g. pilocytic astrocytoma or Lynch syndrome patients (5, 12, 13).
Intron 19 of the ALK gene resembles the breakpoint cluster region and has a size of about 1,932 bp. Thus, the LD-/LDI-PCR seems to be a highly feasable method, as it is for MLL, PML-RARA or FGFR1 rearrangement analyses (3, 4, 9-11). LDI-PCR could be expected to be a powerful method to analyze all EML4-ALK variants and also any other ALK rearrangements having known or even unknown fusion partner genes (Fig. 1). Therefore, we believe that the implementation of such a novel technology for the molecular diagnoses of lung cancer patients would contribute to a more accurate identification of ALK fusion genes in NSCLC as well as to the molecular characterization of EML4-ALK fusion gene in Korean or other ethnic lung cancer patients that can be used as a more precise founding information for molecular targeted therapy. We hope this new LD-/LDI-PCR method can be used in a greater variety of molecular diagnosis of solid tumors in the near future.
Figures and Tables
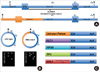
Fig. 1
Schematic representation of long distance inverse-polymerase chain reaction (LDI-PCR) for analyzing ALK rearrangements. LDI-PCR can analyze any kind of EML4-ALK fusion variant and other (or unknown) ALK partner genes. (A) One ALK wildtype allele and one rearranged ALK allele are presented. Genomic breakpoint cluster region (BCR) of ALK in non-small cell lung cancer (NSCLC) is located in 19th intron of ALK gene. (R: restriction enzyme) (B) General principle of LDI-PCR for the detection of ALK fusion gene analysis. Two asterisks show the derivative (target) bands by LDI-PCR in ALK fusion gene analysis. (C) Demonstration of known (EML4, TFG, KIF5B, KLC1) or unknown partner genes in ALK rearrangements.
References
1. Jin G, Jeon HS, Lee EB, Kang HG, Yoo SS, Lee SY, Lee JH, Cha SI, Park TI, Kim CH, et al. EML4-ALK fusion gene in Korean non-small cell lung cancer. J Korean Med Sci. 2012. 27:228–230.
2. Kim H, Yoo SB, Choe JY, Paik JH, Xu X, Nitta H, Zhang W, Grogan TM, Lee CT, Jheon S, et al. Detection of ALK gene rearrangement in non-small cell lung cancer: a comparison of fluorescence in situ hybridization and chromogenic in situ hybridization with correlation of ALK protein expression. J Thorac Oncol. 2011. 6:1359–1366.
3. Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009. 23:1490–1499.
4. Meyer C, Schneider B, Reichel M, Angermueller S, Strehl S, Schnittger S, Schoch C, Jansen MW, van Dongen JJ, Pieters R, et al. Diagnostic tool for the identification of MLL rearrangements including unknown partner genes. Proc Natl Acad Sci U S A. 2005. 102:449–454.
5. Yang JJ, Marschalek R, Meyer C, Park TS. Diagnostic usefulness of genomic breakpoint analysis in various gene rearrangements of acute leukemias: a perspective of LD- or LDI-PCR based approaches. Ann Lab Med. 2012. [In press].
6. Kim MJ, Choi JR, Suh JT, Lee HJ, Lee WI, Park TS. Diagnostic standardization of leukemia fusion gene detection system using multiplex reverse transcriptase-polymerase chain reaction in Korea. J Korean Med Sci. 2011. 26:1399–1400.
7. Sasaki T, Rodig SJ, Chirieac LR, Jänne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010. 46:1773–1780.
8. Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, Nakajima T, Mano H, Takeuchi K. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One. 2012. 7:e31323.
9. Yang JJ, Park TS, Choi JR, Park SJ, Cho SY, Jun KR, Kim HR, Lee JN, Oh SH, Lee S, et al. Submicroscopic deletion of FGFR1 gene is recurrently detected in myeloid and lymphoid neoplasms associated with ZMYM2-FGFR1 rearrangements: a case study. Acta Haematol. 2012. 127:119–123.
10. Lim G, Cho EH, Cho SY, Shin SY, Park JC, Yang YJ, Oh SH, Marschalek R, Meyer C, Park TS. A novel PML-ADAMTS17-RARA gene rearrangement in a patient with pregnancy-related acute promyelocytic leukemia. Leuk Res. 2011. 35:e106–e110.
11. Kim MJ, Cho SY, Kim MH, Lee JJ, Kang SY, Cho EH, Huh J, Yoon HJ, Park TS, Lee WI, et al. FISH-negative cryptic PML-RARA rearrangement detected by long-distance polymerase chain reaction and sequencing analyses: a case study and review of the literature. Cancer Genet Cytogenet. 2010. 203:278–283.
12. Cin H, Meyer C, Herr R, Janzarik WG, Lambert S, Jones DT, Jacob K, Benner A, Witt H, Remke M, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011. 121:763–774.
13. Meyer C, Brieger A, Plotz G, Weber N, Passmann S, Dingermann T, Zeuzem S, Trojan J, Marschalek R. An interstitial deletion at 3p21.3 results in the genetic fusion of MLH1 and ITGA9 in a Lynch syndrome family. Clin Cancer Res. 2009. 15:762–769.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download