Abstract
Panton-Valentine leukocidin (PVL)-positive USA300 clone has been the most successful community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) clone spreading in North America. In contrast, PVL-negative ST72-CA-MRSA has been predominant in Korea, and there has been no report of infections by the USA300 strain except only one case report of perianal infection. Here, we describe the first case of pneumonia caused by the USA300 strain following pandemic influenza A (H1N1) in Korea. A 50-year-old man was admitted with fever and cough and chest radiograph showed pneumonic consolidation at the right lower lung zone. He received a ventilator support because of respiratory failure. PCR for pandemic influenza A (H1N1) in nasopharyngeal swab was positive, and culture of sputum and endotracheal aspirate grew MRSA. Typing of the isolate revealed that it was PVL-positive, ST 8-MRSA-SCCmec type IV. The analysis of the PFGE patterns showed that this isolate was the same pulsotype as the USA300 strain.
During the past decade, community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) has emerged worldwide. Although there have been some evidences of intercontinental spread of major CA-MRSA clones, most CA-MRSA clones showed the continent-specific distribution (1). Especially, Panton-Valentine leukocidin (PVL)-positive USA300 (multilocus sequence type [ST] 8) clone has been reported as the most successful CA-MRSA clone spreading in the community and in hospitals in North America (1). The study of bacterial isolates from purulent skin and soft tissue infections in 11 US emergency departments showed that 59% of all cases of skin and soft tissue infections were caused by MRSA and 97% of them were USA300 (2). A prospective study in San Francisco showed that the USA300 clone accounted for 78.5% of community-onset and 43.3% of hospital-onset MRSA infections indicating that this clone had spread rapidly in the community and also in hospitals in this region.
In contrast, in Korea, PVL-negative ST72-MRSA-SCCmec type IV has been reported as the predominant CA-MRSA strain (3-5), and there has been no report of infections by the USA300 strain except one case report of perianal infection, which was suggested to be imported from Hawaii (6). Here, we describe the first case of pneumonia caused by the USA300 strain following pandemic influenza A (H1N1) in Korea.
A 50-yr-old man, who had been healthy except a history of prostatic cancer two years ago, was admitted to Samsung Medical Center, Seoul, Korea, on November 21, 2009, with high fever and productive cough for three days. He complained of hemoptysis with right pleuritic chest pain. His family members and he had not traveled to other countries in the previous year. On admission, he had fever (38.9℃), pulse rate of 65 beats/min, blood pressure of 184/154 mmHg, and respiratory rate of 20 breaths/min. Physical examination showed decreased breath sounds at the right lower lung field. Laboratory tests showed 6,860 leukocytes/µL (78.6% neutrophils and 15.2% lymphocytes), hemoglobin 14.8 g/dL, and platelet 154,000/µL. Other laboratory values included serum sodium 131 mM/L, blood urea nitrogen 14.1 mg/dL, creatinine 1.2 mg/dL, glucose 138 mg/dL, and lactic acid 2.3 mM/L. Arterial blood gas analysis showed pH 7.483, pCO2 29.9 mmHg, pO2 59.9 mmHg, HCO3 21.9 mM/L and SaO2 91.5% on room air. His chest radiograph showed pneumonic consolidation at the right lower lung zone. Real-time reverse transcription PCR in nasopharyngeal swab was positive for pandemic influenza A (H1N1). Gram stain of sputum showed many Gram-positive cocci and urinary antigen test for Streptococcus pneumoniae was negative. He was given ceftriaxone and levofloxacin empirically as well as oseltamivir. On the same day of admission, his condition rapidly deteriorated and he was transferred to the intensive care unit. He received a ventilator support after intubation because of respiratory failure. A computed tomography scan of the chest showed necrotizing pneumonia involving the right lower lobe (Fig. 1). Culture of sputum and endotracheal aspirate grew MRSA, and it was susceptible to gentamicin, clindamycin, rifampicin, and trimethoprim/sulfamethoxazole, however, resistant to ciprofloxacin, erythromycin, and tetracycline. The D-test for inducible clindamycin resistance was negative. The patient suffered from hemoptysis and substantial amount of purulent sputum over 2 weeks. He was fully recovered after vancomycin treatment for 2 weeks followed by oral rifampicin and trimethoprim/sulfamethoxazole for 3 weeks.
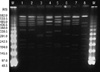
For genotyping of the bacterial isolate, multilocus sequence typing (MLST) was carried out by PCR amplification and sequencing of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) as previously described (7). The allelic profiles and sequence types (STs) were assigned by the MLST web site (http://saureus.mlst.net/). SCCmec types were determined by the multiplex PCR method. Staphylococcal protein A (spa) typing was performed as previously described (8). The spa types were determined by using Ridom SpaServer (http://spaserver2.ridom.de/spatypes.shtml). Isolates were screened for the lukF-PV and lukS-PV genes encoding the components of the PVL toxin by PCR amplification of a portion of both the lukS-PV and lukF-PV ORFs by using the primer pair luk-PV-1 and luk-PV-2 designed by Lina et al. (9). Pulsed field gel electrophoresis (PFGE) was performed as described previously (10). The PFGE patterns were analysed using GelCompar II software (Applied Maths, Belgium). Typing of the MRSA isolate from the patient revealed that the isolate belonged to MLST ST8, carried SCCmec type IV, and was positive for PVL. The spa type was t008 (Ridom Staph Type). The analysis of the PFGE patterns showed that this isolate was the same pulsotype as the USA300 strain, which was obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA), supported under NIAID/NIH contract #HHSN2722 0070 0055C (Fig. 2). The arcA gene in the arginine catabolic mobile element (ACME) which is characteristic for the USA300 strain was also positive.
The USA300 clone has been the most successful CA-MRSA clone epidemiologically during the past decade, and has become widely prevalent strain in the USA. Although there have been increasing reports of the cases of infections by the USA300 clone from some European countries (11) and reports of sporadic cases in Japan (12), it has been considered that transcontinental spread of the USA300 clone has not been successful yet. In Korea, there have been a few surveillance studies characterizing CA-MRSA strains in Korea, however, the USA300 strain was not found (3, 4). There has been only one case report of perianal infection caused by the USA300 strain, which was suggested to be imported from Hawaii (6). This is the first case report of pneumonia caused by the USA300 clone in Korea, which had manifested as a post-influenza A (H1N1) pneumonia.
We demonstrated that the MRSA isolate from our case was the same strain as the USA300 clone using various genotypic methods including MLST, SCCmec typing, spa typing, and PFGE. The fact that his family members and he had not traveled to the USA suggests the possibility that the USA300 clone might have been spreading in Korea, although there has been no report revealing the presence of the carriers of the USA300 strain in Korea to date (13).
Although the USA300 CA-MRSA clone has emerged as a major pathogen causing skin and soft tissue infections, it has also been associated with invasive diseases including necrotizing pneumonia, bacteremia, and osteomyelitis. Furthermore, S. aureus has been reported as one of the important pathogens causing bacterial pneumonia following pandemic influenza A (H1N1) (14) as well as previous pandemic (15) and seasonal influenza (16). Post-mortem study of 77 fatal cases due to pandemic influenza A (H1N1) by the Centers for Disease Control and Prevention (CDC) identified 5 cases of MRSA infection, although genotypic information was not given in this report (14). The cases of coinfection with CA-MRSA and pandemic influenza A (H1N1) have been also reported in other countries including Australia (17). This is important because S. aureus is not a common pathogen causing community-acquired pneumonia in general; however we have to consider S. aureus as a very likely pathogen in influenza season. Furthermore, animal experiments demonstrated that antecedent influenza virus infection had a profound influence on the morbidity and mortality associated with S. aureus pneumonia (18), and clinical data supported this (19).
Early appropriate antimicrobial treatment is very important for treatment of pneumonia caused by CA-MRSA, because these strains are resistant to cephalosporins, most commonly prescribed antibiotics for empirical treatment of community-acquired pneumonia. Previous reports have revealed very high mortality rates in the cases of CA-MRSA pneumonia (20). In a clinician's viewpoint, in cases of community-acquired pneumonia occurring during an influenza season, which show the finding of necrotizing pneumonia, the possibility of CA-MRSA should be considered as one of the potential pathogens and empirical antimicrobial agents targeting CA-MRSA should be selected until culture results are reported. Furthermore, considering the possibility of the spread of the USA300 clone and the increasing incidence of community-associated infections by ST72-MRSA in Korea, continuous surveillance of the changing epidemiology of CA-MRSA in Korea is essential.
Figures and Tables
Fig. 1
Chest computerized tomography (CT) scan shows a dense consolidation and necrotizing areas of the right lower lobe.
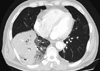
Fig. 2
Pulsed-field gel electrophoresis (PFGE) patterns of MRSA isolate from the patient and control strains obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA). PFGE was performed with restriction enzyme SmaI. Lane 1, isolate from the patient; lane 2, NRS384 (USA300); lane 3, NRS123 (USA400); lane 4, NRS385 (USA500); lane 5, NRS386 (USA700); lane 6, NRS387 (USA800); lane 7, NRS483 (USA1000); lane 8, NRS484 (USA1100); M, lambda marker.
References
1. Deleo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010. 375:1557–1568.
2. Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. EMERGENcy ID Net Study Group. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006. 355:666–674.
3. Kim ES, Song JS, Lee HJ, Choe PG, Park KH, Cho JH, Park WB, Kim SH, Bang JH, Kim DM, Park KU, Shin S, Lee MS, Choi HJ, Kim NJ, Kim EC, Oh MD, Kim HB, Choe KW. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J Antimicrob Chemother. 2007. 60:1108–1114.
4. Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, Yeom JS, Kim SW, Chang HH, Kim YS, Jung SI, Son JS, So TM, Lalitha MK, Yang Y, Huang SG, Wang H, Lu Q, Carlos CC, Perera JA, Chiu CH, Liu JW, Chongthaleong A, Thamlikitkul V, Van PH. ANSORP Study Group. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011. 66:1061–1069.
5. Bae IG, Kim JS, Kim S, Heo ST, Chang C, Lee EY. Genetic correlation of community-associated methicillin-resistant Staphylococcus aureus strains from carriers and from patients with clinical infection in one region of Korea. J Korean Med Sci. 2010. 25:197–202.
6. Park CM, Lee DG, Choi SM, Park SH, Choi JH, Yoo JH, Hur JA, Shin WS. A case of perianal abscess due to Panton-Valentine leukocidin positive community-associated methicillin-resistant Staphylococcus aureus: report in Korea and literature review from the Far East. Infect Chemother. 2008. 40:121–126.
7. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000. 38:1008–1015.
8. Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004. 42:792–799.
9. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999. 29:1128–1132.
10. Yinduo J. Methods in molecular biology: MRSA protocols. 2007. Totowa, NJ: Humana Press, Inc..
11. Blanco R, Tristan A, Ezpeleta G, Larsen AR, Bes M, Etienne J, Cisterna R, Laurent F. Molecular epidemiology of Panton-Valentine leukocidin-positive Staphylococcus aureus in Spain: emergence of the USA300 clone in an autochthonous population. J Clin Microbiol. 2011. 49:433–436.
12. Nagao M, Linuma Y, Suzuki M, Matsushima A, Takakura S, Ito Y, Ichiyama S. First outbreak of methicillin-resistant Staphylococcus aureus USA300 harboring the Panton-Valentine leukocidin genes among Japanese health care workers and hospitalized patients. Am J Infect Control. 2010. 38:e37–e39.
13. Kwon JC, Kim SH, Park SH, Choi SM, Lee DG, Choi JH, Park C, Shin NY, Yoo JH. Molecular epidemiologic analysis of methicillin-resistant Staphylococcus aureus isolates from bacteremia and nasal colonization at 10 intensive care units: multicenter prospective study in Koera. J Korean Med Sci. 2011. 26:604–611.
14. Centers for Disease Control and Prevention (CDC). Bacterial co-infection in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1): United States, May-August 2009. MMWR Morb Mortal Wkly Rep. 2009. 58:1071–1074.
15. Chickering HT, Park JH. Staphylococcus aureus pneumonia. JAMA. 1919. 72:617–626.
16. Pallin DJ, Egan DJ, Pelletier AJ, Espinola JA, Hooper DC, Camargo CA Jr. Increased US emergency department visits for skin and soft tissue infections, and changes in antibiotic choices, during the emergence of community-associated methicillin-resistant Staphylococcus aureus. Ann Emerg Med. 2008. 51:291–298.
17. Murray RJ, Robinson JO, White JN, Hughes F, Coombs GW, Pearson JC, Tan HL, Chidlow G, Williams S, Christiansen KJ, Smith DW. Community-acquired pneumonia due to pandemic A(H1N1)2009 influenzavirus and methicillin resistant Staphylococcus aureus co-infection. PLoS One. 2010. 5:e8705.
18. Lee MH, Arrecubieta C, Martin FJ, Prince A, Borczuk AC, Lowy FD. A postinfluenza model of Staphylococcus aureus pneumonia. J Infect Dis. 2010. 201:508–515.
19. Hageman JC, Uyeki TM, Francis JS, Jernigan DB, Wheeler JG, Bridges CB, Barenkamp SJ, Sievert DM, Srinivasan A, Doherty MC, McDougal LK, Killgore GE, Lopatin UA, Coffman R, MacDonald JK, McAllister SK, Fosheim GE, Patel JB, McDonald LC. Severe community-acquired pneumonia due to Staphylococcus aureus, 2003-04 influenza season. Emerg Infect Dis. 2006. 12:894–899.
20. Boussaud V, Parrot A, Mayaud C, Wislez M, Antoine M, Picard C, Delisle F, Etienne J, Cadranel J. Life-threatening hemoptysis in adults with community-acquired pneumonia due to Panton-Valentine leukocidin-secreting Staphylococcus aureus. Intensive Care Med. 2003. 29:1840–1843.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download