Abstract
Long-term synaptic plasticity requires addition of new proteins at the synaptic site. The local protein synthesis at subsynaptic sites confers advantageous mechanisms that would regulate the protein composition in local domains on a moment-by-moment basis. However, our information on the identities of 'dendritic' mRNAs is very limited. In this study we investigated the expression of the protein and mRNA for eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4EBP1) in cultured rat hippocampal neurons. Immunocytochemistry (ICC) showed that 4EBP1 protein is highly localized to the nucleus. In dendrites most 4EBP1 punctae were not colocalized with those of eIF4E. In situ hybridization (ISH) and Fluorescence ISH (FISH) revealed that 4EBP1 mRNA was present in dendrites. The FISH signals formed clusters along dendrites that colocalized with ICC signals for Staufen, a marker for RNA granules. The neuronal activation by KCl (60 mM, 10 min) significantly increased the density of 4EBP1 FISH signals in the nucleus after 2 hr, and both in the nucleus and dendrites after 6 hr. Our results indicate that 4EBP1 and its mRNA are present in dendrites, and the mRNA is upregulated and transported to dendritic domains in RNA granules upon neuronal activation.
Neuronal synapses continuously change their efficacy of neurotransmission depending on the history of usage. This so-called synaptic plasticity is thought to underlie the cellular basis for learning and memory (1). Although short-term synaptic plasticity occurs by modifications of existing proteins (2), long-term plasticity such as long-term potentiation and long-term depression requires addition of new proteins at the synaptic site [see a recent review by Skup (3)]. Translation in neurons primarily occurs in the soma. However, recent studies show that local protein synthesis at synapses can be triggered by synaptic activation. The on-site synthesis of protein within neuronal dendrites confers advantageous mechanisms that would regulate the protein composition in local domains on a moment-by-moment basis. Dendritic protein synthesis was first suggested by the discovery of polyribosomes and membranous cisterns beneath postsynaptic sites (4, 5). Thereafter, other components of the translation machinery were identified, and dendritic mRNAs were identified by various experimental techniques (3). The local protein synthesis was finally proved in dendrites which were mechanically separated from neuronal perikarya (6).
The cytoplasmic granules containing translationally repressed mRNAs are present in germ cells, embryos and neurons, and they are collectively called RNA granules (7). A subset of mRNAs is localized to the dendrites (3, 8, 9). The dendritic mRNAs relocalize to the activated postsynaptic sites (10) and protein synthesis is localized to "hot spots" that occur in proximity to synapses (6). RNA granules are large tightly clustered aggregates of ribosomes (7). They also contain some components of translation, including ribosomal RNA, mRNAs, and translation factor such as elongation factor 1A (eEF1A) (7, 11). However, the rate-limiting factors in translation initiation (eIF) such as eIF4E and 4G are present only in trace amounts in the RNA granules (12). Therefore, it is unlikely that Cap-dependent or Cap-independent initiation occurs in the granules (13). Where and how the missing translation factors are implemented? Synapse-specific plasticity of synaptic transmission suggests that the missing factors are supplied on-site in the subsynaptic area. For example, eIF4E is mainly concentrated in the dendritic lipid raft in close proximity to the postsynaptic sites (14), and treatment of various neuronal preparations with the brain-derived neurotrophic factor results in redistribution of the eIF4E into dendritic spines (15). Other translation factors eIFs (16) and eEF1A (17) and are enriched in the postsynaptic sites.
In addition to the translation factors our laboratory reported that their mRNAs are also present at the postsynaptic site. The translation initiation factor eIF4E (18) and elongation factor eEF1A mRNAs (19) are localized to neuronal dendrites, and neuronal activation by KCl causes their upr egulation and relocalization to subsynaptic sites. In the present report we provide evidence that eIF4E-binding protein 1 (4EBP1) and its mRNA are also localized to dendrites in unstimulated rat hippocampal neurons, and that KCl treatment upregulates the mRNA in dendrites.
Dissociated hippocampal cells from Sprague-Dawley rat pups at embryonic day 18 (E18) or E19 were plated onto 12-mm diameter polylysine/laminin-coated glass coverslips at a density of 150 neurons/mm2 as described (20) and grown in astrocyte-conditioned Neurobasal media as described (21, 22). Cells were fixed by a sequential paraformaldehyde (PFA)/methanol (MeOH) fixation procedure (incubation in 4% PPA in PBS [20 mM sodium phosphate buffer, pH 7.4, 0.9% NaCl] at room temperature for 10 min followed by incubation in MeOH at -20℃ for 20 min) (22). For KCl treatment coverslips were transferred to a new medium containing KCl (60 mM) and incubated for 10 min. Coverslips were rinsed in a fresh medium without KCl and returned to the original culture wells. Cells were fixed after further incubation for 2 or 6 hr and subjected to FISH.
A 353 bp DNA fragment encoding the rat eIF4EPB1 gene (ref-|NM_053857.1) mRNA (-21 ~ +403; this covers the full-length [aa. 1~117] of the structural gene) was amplified from a cDNA library (Marathon-Ready™ cDNA , Clontech, Mountain View, CA, USA) by polymerase chain reaction (PCR) with the primers: sense 5'-ccAGATCTcgtgcaggag acatgtc-3'/antisense: 5'-gcGGTACCaatgtccatctcaaactgtg-3'. Capital letters indicate sequences of restriction sites (BglII and KpnI, respectively) added for cloning purposes. The PCR product was cloned into the BglII and KpnI sites of a pBluescript II SK(-) transcription vector (Invitrogen). The digoxigenin (DIG)-labeled antisense and sense riboprobes were prepared by in vitro transcription of the linearlized plasmids (BamHI for antisence, EcoRI for sense) with SP6 or T7 RNA polymerase using DIG-RNA Labeling mix according to manufacturer's instruction (Roche Applied Science, Indianapolis, IN, USA).
The PFA/MeOH fixed cells were incubated overnight with DIG-labeled riboprobes (200 ng/mL) at 53℃ in 400 µL of the NorthernMax Prehybridization/Hybridization buffer (Ambion, Austin, TX, USA) and washed finally in 0.1X SSC at 53℃ for 15 min. The riboprobes were visualized by color development using monoclonal biotin-conjugated anti-digoxin antibodies (Clone DI-22, Sigma, St. Louis, MO, USA), 1-Step NBT/BCIP substrate (Pierce, Rockford, IL, USA), and alkaline phosphatase-conjugated streptavidin (diluted 1:1,000; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) as described (22).
Combined FISH and ICC were performed with primary (biotin-conjugated mouse monoclonal anti-digoxin [1:500; Sigma], rabbit anti-4EBP1 [R-113, 1:200; Santa Cruz Biotech., Heidelberg, Germany], chicken anti-PSD-95 [1:1,000; antiserum UCT-C1, Murphy et al. (23), or rabbit anti-staufen [1:100; Chemicon]) and secondary antibodies (Alexa Fluor 488-conjugated Streptavidin, Alexa Fluor 488-conjugate goat anti-mouse, 568-conjugated goat anti-rabbit, and Alexa Fluor 647-conjugated goat anti-chicken IgG [each diluted 1:1,000 in blocking buffer; Invitrogen]) as described (22).
A Leica Research Microscope DM IRE2 (Leica Microsystems AG, Wetzlar, Germany) was used for light microscopy of color-developed ISH samples. Images were acquired with a high-resolution CoolSNAP™ CCD camera (Photometrics Inc., Germany) under the control of a computer equipped with Leica FW4000 software. Exposure times were set so that pixel brightness was not saturated and were held constant during acquisition of all images (1,388 × 1,039 pixels) for each pair of sense- and antisense-probed cells. Confocal images of combined FISH and ICC (1,024 × 1,024 pixels) were acquired using 100X oil-immersion lens on the Leica TCS SP2 confocal system with laser lines at 488, 543, and 633 nm and processed with the use of Adobe Systems Photoshop 7.0 software.
Densitometric analyses and surface plots were performed with an Image J (version 1.45) software (National Institute of Health, Bethesda, MD, USA). Statistical significance was assessed by Mann-Whitney U-test. The P values less than 0.05 and 0.01 were considered to be significant and very significant, respectively.
To investigate the subcellular localization of 4EBP1 relative to eIF4E, we double-labeled mature hippocampal neurons (16 days in vitro; DIV 16) with α-eIF4E and α-4EBP1. Confocal micrographs of the ICC showed that 4EBP1 was distributed throughout the neuron with highest expression in the nucleus (Fig. 1A, asterisk). In the perikaryon and dendrites, 4EBP1-immunoreactivity (IR) formed small punctae, of which density was slightly lower in dendrites than the perikaryon. This expression profile was very different from that of eIF4E, of which expression level was much higher in the perikaryon than in the dendrites (Fig. 1B, asterisk). The merge image contrasts the relative subcellular localization of the two proteins (Fig. 1C). To see the relative positions of the two proteins, a portion of the dendrite (box) was enlarged and shown in the insets. It can be seen that both eIF4E (green) and 4EBP1 (red) punctae were densely distributed in the dendritic shaft (Fig. 1C, insets eIF4E and 4EBP1). Despite of high density in the dendrite the two kinds of punctae rarely overlap each other (Fig. 1C, inset merge).
Local dendritic translation of mRNAs can implement the necessary proteins in the dendrite on a moment-to-moment basis. To investigate if 4EBP1 mRNA is localized to dendrites, we performed ISH of cultured rat hippocampal neurons. After PFA/MeOH fixation, cells were hybridized in situ with DIG-labeled sense (S)- or antisense (AS)-4EBP1 riboprobes, and ISH signals were visualized with biotin-conjugated anti-digoxin antibody and alkaline phosphatase-conjugated streptavidin as detailed in Materials and Methods. As shown in the bright field light micrographs (Fig. 2A), the AS-riboprobe reveals significant ISH signals for 4EBP1 mRNA in dendrites (Fig. 2A, AS-4EBP1, inset, arrowhead), although the strongest signals are associated with the soma (Fig. 2A, AS-PB1, inset, arrow). In contrast, the ISH signal by the sense probe is not significant above background (Fig. 2A, S-4EBP1). We performed ISH for eEF1A mRNA as a general control of the protocol, because we previously showed its localization in both neuronal soma and dendrites (19). The AS-eEF1A riboprobe produces ISH signals in dendrites as well as in the soma (Fig. 2A, AS-eEF1A, arrowhead and arrow, respectively), while the control ISH signal by the S-eEF1A riboprobe is negligible (Fig. 2A, S-eEF1A). This result indicates that the general process of the ISH is integral, and that the ISH signals for 4EBP1 mRNA are reliable.
To verify that the signals in dendrites shown by ISH represented RNA granules, we performed FISH combined with ICC to show colocalization of 4EBP1 FISH signals with staufen protein, a marker protein for RNA granules. PFA/MeOH-fixed hippocampal neurons were subjected to FISH with DIG-labeled 4EBP1 riboprobes, followed by ICC with antibodies against staufen (Fig. 2B). The FISH signals for 4EBP1 mRNA are distributed in clusters along dendrites (Fig. 2B, AS-4EBP1). The ICC signals of staufen are also present as clusters along dendritic shafts (Fig. 2B, α-Stau), a distribution profile similar to previous reports (18, 25). The merged images exhibit that most (86.7±8.4%, n=158) 4EBP1 FISH signals overlap with those of staufen ICC (Fig. 2B, merge, yellow arrowheads). This result clearly shows that the FISH signals represent RNA granules.
Strategic localization at subsynaptic sites may be important for tight regulation of local translation. Using a sequential PFA/MeOH fixation procedure that produces high signals for both FISH and ICC (22) we investigated the positioning of 4EBP1 mRNA relative to postsynaptic sites. For this purpose we used an antibody against PSD-95 as a postsynaptic marker. The AS-4EBP1 riboprobe reveals FISH signals in clusters along dendrites and soma as well (Fig. 3A, arrowhead and asterisk, respectively). To visualize better the signal intensity a surface plot of the boxed area in the AS-4EBP1 FISH image was done. As shown in Fig. 3B the FISH signal intensity is slightly higher in the nucleus (Nuc) than in the dendrites (Den). The ICC signals for PSD-95 are robust and they are highly concentrated in clusters along dendrites (Fig. 3A, α-PSD95). A merge image is shown in Fig. 3A (merge), where the boxed area is shown enlarged in the inset. In the single channel of Fig. 3A inset images it can be seen that the 4EBP1 FISH signals (green) are randomly distributed in the dendritic shaft. This spatial distribution was in contrast to that of PSD-95 protein (red): The PSD-95 clusters were positioned toward the periphery of the shaft. The merge image shows that the two signals rarely overlap, indicating that 4EBP1 mRNAs are not specifically localized to the postsynaptic sites.
To investigate whether the dendritic 4EBP1 mRNA was regulated by neuronal activity, we added 60 mM KCl to the culture medium for 10 min to depolarize the neurons. Neurons were fixed 2 or 6 hr later and processed for FISH combined with ICC. The confocal micrographs of 2hr treatment are shown in Fig. 4A. The FISH signal intensity for 4EBP1 mRNA in the nucleus is significantly (67%, P < 0.01) higher (Fig. 4A, AS-4EBP1, arrow; Fig. 4D) than in the control (Fig. 3A, AS-4EBP1). In this particular microscopic field there are many PSD-95-negative non-neuronal cells, which also show significant FISH signals in the nucleus (arrowhead). In contrast to the nucleus, the increase in the dendrite was not significant. To visualize better the expression profile, a surface plot of the boxed area was done. As shown in Fig. 4C, 2 hr, the signal intensity in the nucleus is very significantly (P < 0.01; Fig. 4D) increased compared to the control (Fig. 3A, AS-4EBP1). The sense 4EBP1 riboprobe (s-4EBP1) does not produce significant FISH signals (Fig. 4A, S-4EBP1). However, there appears a couple of large clusters in the nucleus of both neuronal (arrow) and non-neuronal cells (arrowhead). Similar clusters are also seen in the cells treated with KCl for 6 hr. It is likely that this nucleoplasmic structure contains nucleotide sequences that had homology to s-4EBP1.
Typical confocal images of 4EBP1 FISH after 6 hr of KCl treatment is shown in Fig. 4B. The 4EBP1 FISH signal in the nucleus is dramatically increased 6 hr after stimulation compared to unstimulated or 2 hr-stimulated neurons (Fig. 4B, AS-4EBP1; asterisk in inset). The increase in the signal intensity is visualized in a surface plot of the boxed area (Fig. 4C, 6 hr). Statistical analyses showed that, compared to the untreated control and 2 hr-stimulated, the FISH signal intensity for 4EBP1 mRNA in the nucleus increases by 3-fold and 2-fold, respectively (Fig. 4D). At this time of stimulation, the increase in the 4EBP1 FISH signal intensity is also evident in the dendrite (Fig. 4B, AS-4EBP1 and Fig. 4C, 6 hr). Statistical analysis showed that, compared to both unstimulated and 2 hr-stimulation, the intensity increased very significantly (45%, P < 0.01) (Fig. 4D). These data show that 4EBP1 mRNA was upregulated by the KCl treatment, and that stimulation for 6 hr was long enough for the newly transcribed mRNA was transported to the dendrite.
Our ICC data also show that PSD-95 clusters are significantly upregulated by KCl stimulation in the dendrites of hippocampal neurons: 50% increase at both 2 and 6 hr after KCl treatment; from 14.6±2.5 (control) to 22.0±3.3 (2 hr) and 21.3±3.7 (6 hr) per 30 µm. Since previous studies had shown that PSD-95 was upregulated both in vivo and in vitro by neuronal activation (27-29), this result verifies that the neurons were adequately stimulated in our experimental procedures.
In this study we have shown that differential distribution of the two proteins eIF4E and its binding protein 4EBP1 in cultured hippocampal neurons, and that 4EBP1 mRNA is upregulated in the dendrites by KCl stimulation. Although eIF4E and its binding protein 4EBP1 are functionally related to each other, our ICC images of cultured hippocampal neurons showed difference in the subcellular distribution. 4EBP1 was distributed throughout the neuron with highest expression in the nucleus, whereas the expression level of eIF4E was much higher in the perikaryon than the dendrites. Subtle differential distribution of the two proteins was also seen in the perikaryon and dendrites. In these subcellular domains both eIF4E and 4EBP1 formed small punctae. Despite high density, however, the two kinds of punctae rarely overlap each other, indicating that most eIF4Es in dendrites are not under inhibition. Rong et al. (24) reported that a sizable fraction (~30%) of 4EBP1 is localized to the nucleus in flow cytometric analysis of mouse embryo fibroblasts (MEFs). High nuclear localization of 4EBP1 was also proved by ICC of MEFs and immunohistochemistry of the mouse kidney. They further showed that, in serum-starved and/or rapamycin-treated MEFs, nuclear 4EBP1 sequester eIF4E in the nucleus, indicating that 4EBP1 is a regulator of eIF4E cellular localization. In this study we also found that 4EBP1 is highly localized in the neuronal nucleus. Neuronal nuclear 4EBP1 may sequester eIF4E in analogy to MEFs.
Our present study has shown that 4EBP1 mRNA is present and can be upregulated in dendrites by neuronal activation. Local dendritic translation of mRNAs can implement the necessary proteins in the dendrite on a moment-to-moment basis. The presence of 4EBP1 mRNA was firstly demonstrated by ISH, and the identity of the RNA granule was further verified by colocalization of 4EBP1 FISH signals with staufen protein ICC signals. Strategic localization at subsynaptic sites may be important for tight regulation of local translation. Most of the 4EBP1 mRNA FISH signals did not overlap with PSD95 ICC signals. Since PSD-95 clusters are positioned toward the periphery of the shaft due to their localization at the spine heads (26), our data indicate that 4EBP1 mRNAs are not specifically localized to the postsynaptic sites. This result is in contrast to the distribution of mRNAs for other translation factors. We previously reported that more than half of eIF4E and eEF1A mRNA FISH clusters overlap with or are immediately adjacent to PSD-95 ICC clusters (18, 19). The differential distribution of 4EBP1 is also shown at the protein level: In contrast to the high concentration at the subsynaptic sites, the 4EBP1 proteins were distributed throughout the shaft of dendrites (this study). For example, eEF1A were highly enriched at the postsynaptic sites (26). Translation initiation factors such as eIF4E and eIF4G (which bind the 5'-terminal mRNA cap), eIF5 (which is important during the 3' direction scanning to find an initiation codon), eIF6 (which mediates upregulation of translation by external stimuli), and eIF5A (which mediate translation upregulation under adverse conditions) were localized to the postsynaptic sites (16).
Why is 4EBP1 not localized at the subsynaptic site? The reason for this contrast is not clear. However, the difference could originate from the different roles in translation. In contrast to the positive role of other translation factors, the 4EBP1 is a negative regulator in translation. It is reasonable to conceive that positive translation factors should be readily available at a moment-to-moment basis upon synaptic activation, whereas a negative regulator could be implemented later in time after the necessary protein neosynthesis is completed. This temporal difference in the necessity may be represented, in turn, in the differential spatial distribution in dendrites.
The present study further shows that the dendritic 4EBP1 mRNA is upregulated by neuronal activation. Our data show that 4EBP1 mRNA is upregulated by the KCl treatment, and that stimulation for 6 hr is long enough for the newly transcribed mRNA is transported to the dendrite. Our ICC data also show that PSD-95 clusters are significantly upregulated by KCl. Since previous studies had shown that PSD-95 was upregulated both in vivo and in vitro by neuronal activation (27-29), this result verifies that the neurons were adequately stimulated in our experimental procedures.
Local protein synthesis in the neuronal dendrite is a critical component of synaptic plasticity. In long-lasting hippocampal synaptic plasticity and memory, mammalian target of rapamycin (mTOR) serves as one of the primary triggers for the initiation of cap-dependent translation via phosphorylation of eIF4EBPs and S6Ks (30). Unphosphorylated 4EBP binds tightly to eIF4E, whereas 4EBP phosphorylated by mTOR does not, thereby permitting initiation to proceed (31). Phosphorylated S6K, in turn, phosphorylates downstream targets such as ribosomal protein S6 and eIF4B (32). Previously we reported that the eIF4E mRNA is localized, and upregulated by neuronal activation, near postsynaptic sites (18). The presence, and upregulation, of its regulatory protein 4EBP1 in dendrites could be a necessary sequel to the increase in eIF4E. Recently, Qi et al. (33) reported that mTOR was activated in hippocampus of the learning rat. The substrates of mTOR, 4EBP and p70S6K were phosphorylated, and total protein synthesis was enhanced, in the learning hippocampus. Their results show that mTOR signaling pathways are activated and the general translation is enhanced in a learning paradigm. In contrast to our in vitro study, the amounts of these proteins were not changed in their in vivo learning paradigm. The discrepancy could originate from the extent of neuronal activation. To support this assumption the mTOR and p70S6K phosphorylation was not increased in either cerebral cortices or cerebellums, which would be less activated during the learning paradigm. Therefore, it would be interesting to carry out animal study if mRNAs for translation factors are upregulated in the learning brain.
Figures and Tables
Fig. 1
Expression of 4EBP1 and eIF4E. Confocal micrographs of dissociate rat hippocampal neurons (DIV16) double-labeled with 4EBP1 (A) and eIF4E (B). A merge image (C) is shown and the insets show the boxed area enlarged in single channels. The boxed area in the inset was further enlarged on the right. The punctae with overlapping signals are marked with arrowheads. * in A, nucleus. * in B, perikaryon. Scale bar, 75 µm.
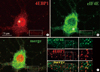
Fig. 2
ISH and FISH showing dendritic localization of 4EBP1 mRNA. (A) ISH. DIV16 dissociated rat hippocampal neurons were hybridized with DIG-labeled antisense (a) or sense (b) cRNA probes prepared by in vitro transcription. The boxed area was shown enlarged in the inset. Sister hippocampal neurons were hybridized with DIG-labeled antisense (c) or sense (d) eIF4E cRNA probes. The soma and dendrites are marked by an arrow and an arrowhead, respectively. (B) Colocalization of FISH signals with staufen immunopuncta. Sister hippocampal neurons were hybridized with DIG-labeled anti-sense 4EBP1 cRNA probes (a) followed by ICC with antibodies against staufen (b). The 4EBP1 mRNA clusters colocalizing with staufen immunopuncta were marked by arrowheads in single (a, b) and merge images (c). Scale bars; 75 µm.
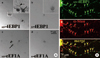
Fig. 3
Subcellular localization of 4EBP1 mRNA. DIV16 dissociated rat hippocampal neurons were hybridized with the DIG-labeled antisense 4EBP1 cRNA probe followed by ICC with antibodies against PSD-95. Confocal micrographs of FISH (A) and ICC (B) images are shown along with a merge (C). The soma and dendrite are marked with an asterisk and an arrowhead, respectively, in A. The inset in C shows the boxed area enlarged in single channels or merge. A FISH signal colocalizing with the ICC signal is marked with an arrowhead. A surface plot of the boxed area in A is shown in D. The positions of nucleus (Nuc) and dendrite (Den) are marked. Scale bars; 75 µm.
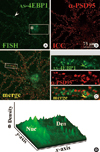
Fig. 4
Upregulation of 4EBP1 mRNA clusters in dendrites of rat hippocampal neurons by KCl treatment. (A) 2 hr post-KCl exposure. Dissociated rat hippocampal cultures (DIV16) were treated with KCl (60 mM, 10 min). After 2 hr, cells were hybridized with DIG-labeled anti-sense 4EBP1 (AS-4EBP1) or sense (S-4EBP1) cRNA probes (FISH) followed by ICC with antibodies against PSD-95 (α-PSD95). Confocal images of single channels or merge are shown. The neuronal and non-neuronal cells are marked with an arrow and an arrowhead, respectively. (B) 6 hr post-KCl exposure. Presentation and markings are similar to A. Neuronal soma and nucleus are marked with an arrow and asterisk, respectively. The large clusters in the neuronal and non-neuronal nuclei (arrow and arrowhead, respectively) are marked in the S-4EBP1 panel. (C) Surface plots. The surface plots of the boxed areas in A and B are shown. The positions of neuronal (Neu-S) and glial (Gli-S) soma are indicated, along with dendrites (Den). (D) Statistics. Statistical significance was assessed by Mann-Whitney U-test. *P < 0.01. Scale bars; 75 µm.
References
1. Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993. 361:31–39.
2. Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term memory - a molecular framework. Nature. 1986. 322:419–422.
3. Skup M. Dendrites as separate compartment - local protein synthesis. Acta Neurobiol Exp (Wars). 2008. 68:305–321.
4. Steward O, Fass B. Polyribosomes associated with dendritic spines in the denervated dentate gyrus: evidence for local regulation of protein synthesis during reinnervation. Prog Brain Res. 1983. 58:131–136.
5. Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982. 2:284–291.
6. Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman E. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001. 30:489–502.
7. Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GH, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996. 16:7812–7820.
8. Tian QB, Nakayama K, Okano A, Suzuki T. Identification of mRNAs localizing in the postsynaptic region. Brain Res Mol Brain Res. 1999. 72:147–157.
9. Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003. 40:347–359.
10. Håvik B, Røkke H, Bårdsen K, Davanger S, Bramham CR. Bursts of high-frequency stimulation trigger rapid delivery of pre-existing alpha-CaMKII mRNA to synapses: a mechanism in dendritic protein synthesis during long-term potentiation in adult awake rats. Eur J Neurosci. 2003. 17:2679–2689.
11. Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995. 108:2781–2790.
12. Krichevsk AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001. 32:683–696.
13. Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011. 23:324–334.
14. Asaki C, Usuda N, Nakazawa A, Kametani K, Suzuki T. Localization of translational components at the ultramicroscopic level at postsynaptic sites of the rat brain. Brain Res. 2003. 972:168–176.
15. Smart FM, Edelman GM, Vanderklish PW. BDNF induces translocation of initiation factor 4E to mRNA granules: evidence for a role of synaptic microfilaments and integrins. Proc Natl Acad Sci USA. 2003. 100:14403–14408.
16. Choi MK, Park SD, Park IS, Moon IS. Localization of translation initiation factors to the postsynaptic sites. J Life Sci. 2011. 21:1526–1531.
17. Cho SJ, Jung JS, Ko BH, Jin I, Moon IS. Presence of translation elongation factor-1A (eEF1A) in the excitatory postsynaptic density of rat cerebral cortex. Neurosci Lett. 2004. 366:29–33.
18. Moon IS, Cho SJ, Seog DH, Walikonis R. Neuronal activation increases the density of eukaryotic translation initiation factor 4E mRNA clusters in dendrites of cultured hippocampal neurons. Exp Mol Med. 2009. 41:601–610.
19. Moon IS, Cho SJ, Lee H, Seog DH, Jung YW, Jin I, Walikonis R. Upregulation by KCl treatment of eukaryotic translation elongation factor 1A (eEF1A) mRNA in the dendrites of cultured rat hippocampal neurons. Mol Cells. 2008. 25:538–544.
20. Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993. 35:567–576.
21. Goslin K, Assmussen H, Banker G. Banker G, Goslin K, editors. Rat hippocampal neurons in low density culture. Culturing Nerve Cells. 1998. 2nd Ed. Cambridge, MA: MIT Press;339–370.
22. Moon IS, Cho SJ, Jin I, Walikonis R. A simple method for combined fluorescence in situ hybridization and immunocytochemistry. Mol Cells. 2007. 24:76–82.
23. Murphy JA, Jensen ON, Walikonis RS. BRAG1, a Sec7 domain-containing protein, is a component of the postsynaptic density of excitatory synapses. Brain Res. 2007. 1120:35–45.
24. Rong L, Livingstone M, Sukarieh R, Petroulakis E, Gingras AC, Crosby K, Smith B, Polakiewicz RD, Pelletier J, Ferraiuolo MA, et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA. 2008. 14:1318–1327.
25. Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999. 19:288–297.
26. Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992. 9:929–942.
27. Skibinska A, Lech M, Kossut M. PSD95 protein level rises in murine somatosensory cortex after sensory training. Neuroreport. 2001. 12:2907–2910.
28. Bao J, Lin H, Ouyang Y, Lei D, Osman A, Kim TW, Mei L, Dai P, Ohlemiller KK, Ambron RT. Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat Neurosci. 2004. 7:1250–1258.
29. Yoshii A, Constantine-Paton M. BDNF induces transport of PSD-95 to dendrites through PI3K-AKT signaling after NMDA receptor activation. Nat Neurosci. 2007. 10:702–711.
30. Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005. 433:477–480.
31. Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001. 15:807–826.
32. Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004. 23:1761–1769.
33. Qi S, Mizuno M, Yonezawa K, Nawa H, Takei N. Activation of mammalian target of rapamycin signaling in spatial learning. Neurosci Res. 2010. 68:88–93.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download