Abstract
This study investigated the spectrum of chromosomal abnormalities in 325 leukemia patients and developed optimal profiles of leukemic fusion genes for multiplex RT-PCR. We prospectively analyzed blood and bone marrow specimens of patients with acute leukemia. Twenty types of chromosomal abnormalities were detected in 42% from all patients by commercially available multiplex RT-PCR for detecting 28 fusion genes and in 35% by cytogenetic analysis including FISH analysis. The most common cytogenetic aberrations in acute myeloid leukemia patients was PML/PARA, followed by AML1/MGT8 and MLL1, and in acute lymphoid leukemia patients was BCR/ABL, followed by TEL/AML1 and MLL1 gene rearrangement. Among the negative results for multiplex RT-PCR, clinically significant t(3;3)(q21;q26.2), t(8;14)(q24;q32) and i(17)(q10) were detected by conventional cytogenetics. The spectrum and frequency of chromosomal abnormalities in our leukemia patients are differed from previous studies, and may offer optimal profiles of leukemic fusion genes for the development of new molecular detection systems.
Cytogenetic aberrations in hematologic malignancies can be assayed by a variety of methods. Commonly used methods are conventional cytogenetics, fluorescence in situ hybridization (FISH), and multiplex reverse transcriptase polymerase chain reaction (RT-PCR) systems. Especially, Pallisgaard et al. have presented a multiplex RT-PCR system that allows simultaneous detection of 29 fusion genes and more than 80 breakpoints and splice variants in patients with acute leukemia (1). Since then, a commercially available multiplex RT-PCR system (HemaVision, DNA technology, Aarhus, Denmark), a similar modified assay, to detect 28 fusion genes in patients with acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL) has been developed.
Chromosomal abnormalities occur in approximately 56% of de novo AML in adults and slightly more frequently in children (2-4). At present, more than 50 different consistently occurring translocations have been described, many of which have been found to be specific for particular subtypes of leukemia or lymphoma (1). To date, most of the data reported were from western countries. Racial differences in various hematological malignancies were previously reported between Asian and western countries (5). It is important, therefore, to investigate and characterize the karyotypic pattern in any geographically restricted population of patients, so that identification of cytogenetic risk factors can be used in current risk-adapted chemotherapy for acute leukemia.
The current study was undertaken to investigate spectra of chromosomal aberrations and diagnostic values of recently developed commercial multiplex RT-PCR system in 325 leukemia patients, and then to recommend a panel of leukemic fusion genes suitable for the development of new efficient molecular detection systems.
We examined 325 leukemia patients during a 4-yr period (2006-2009). Eighty-one children (median age 8 yr, range 0-18 yr) and 244 adults (median age 56 yr, range 19-86 yr) were included in the study. Of the 81 children, 35 (43%) were male and 46 (57%) were female; of the 244 adults, 123 (50%) were male. A total of 213 (66%) were diagnosed as de novo (92%) or secondary (8%) AML (23 children, 190 adults); 104 (32%) were of ALL (55 children, 49 adults); and 8 were of mixed phenotype acute leukemia (MPAL) (Table 1).
Chromosome analysis was performed after short-term culturing without mitogens. The chromosome aberrations were described according to the International System for Cytogenetic Nomenclature 2005 and 2009. A patient was classified as having a normal karyotype only after 20 normal metaphases were analyzed (Fig. 1).
RNA was extracted from peripheral blood or bone marrow using an RNAqueous Kit (Ambion, Austin, TX, USA) according to the manufacturer's protocol (Supplemental Fig. 1). The HemaVision multiplex RT-PCR Screen Test kit (DNA Technology, Aarhus, Denmark) detects 28 of the most common leukemic fusion genes and more than 80 splice variants. In brief, reverse transcription was performed with a mixture of translocation-specific primers. PCR amplification was performed in 2 steps: a master PCR amplification followed by nested PCR which screened for the presence of fusion transcripts and a split-out PCR amplification followed by nested PCR which identified the specific fusion transcript(s). Each of the 8 parallel nested multiplex master PCR reactions contained a mixture of primer pairs for the detection of several fusion transcripts and 2 primer pairs for an internal control gene product of 911 base pairs (Fig. 1). When the presence of one or more fusion transcripts was detected by one or more master PCR reactions, the corresponding split-out reactions with individual primer pairs were performed.
In additional to conventional cytogenetic analysis, FISH was applied in appropriate bone marrow or peripheral blood specimens available from 275 patients. FISH was performed with the Vysis LSI probe (Abbott Molecular/Vysis, Des Plaines, IL, USA) according to the manufacturer's instructions.
Twenty types of chromosomal aberrations in 28 fusion genes were detected in 42% of the total patients by multiplex RT-PCR system (Table 2). The multiplex RT-PCR analysis detected fusion transcripts in 39% (83/213) of the AML patients and in 50% of the ALL patients, in 50% of the MPAL patients (Table 3).
Within the group of 213 AML patients, the following fusion transcripts were detected: 35 PML/PARA, 31 AML1/MGT8, 7 MLL1 rearrangement, 3 CBFB/MYH11, 2 SET/CAN, one of each BCR/ABL, TEL/ABL, AML1/MDS1, PLZF/RARA, TEL/MN1 and TLS/ERG. One child patient with acute leukemic conversion from chronic myeloid leukemia (CML) had simultaneously two fusion transcripts (BCR/ABL and SET/CAN).
In the group of 104 ALL patients, fusion transcripts were detected in 21 BCR/ABL, 15 TEL/AML1, 9 MLL1 including 5 MLL1/AF4, 2 MLL1/AF6, one of each of MLL1/AF9 and MLL1/ENL, 4 E2A/PBX, one of each of TEL/ABL, TEL/ABL and SIL1/TAL1. In the eight patients of the MPAL group, the fusion transcripts were detected in 3 BCR/ABL and one MLL1/ELL (Table 3).
In the group of children, 53% (43/81) exhibited a positive multiplex RT-PCR analysis. Within group of 23 childhood AML patients, the common cytogenetic abnormalities were 7 AML1/MGT8, 4 PML/RARA, one TEL/ABL and one patient with both BCR/ABL and SET/CAN. The major cytogenetic abnormalities in the group of childhood ALL patients (n = 55) were 14 TEL/AML1, followed by 4 BCR/ABL, 4 E2A/PBX and 3 MLL1/AF4.
In the group of adult patients, 39% (96/244) had an abnormal multiplex RT-PCR analysis. Within the group of 190 adult AML patients, the common cytogenetic abnormalities were PML/RARA (n = 31), followed by AML1/MGT8 (n = 24), CBFB/MYH11 (n = 3), MLL1/ENL (n = 3) and MLL1/AF6 (n = 2). The common cytogenetic abnormalities in the adult ALL patients (n = 49) were BCR/ABL (n = 17) and MLL1/AF4 (n = 2) (Supplemental Table 1).
In total, cytogenetic analysis was available in 99% of the AML patients, 99% of the ALL patients and 100% of MPAL. Of the cases, 97% of AML, 89% of ALL and 100% of MPAL were successfully analyzed. Successful cytogenetic analyses were achieved in 307 (94%) patients, among whom 148 (48%) had detectable clonal abnormalities, whereas 159 (52%) were considered cytogenetically normal.
Fourteen types in 28 fusion genes were detected in 35% (113/325) by conventional cytogenetic analysis including FISH. The most frequent cytogenetic abnormality was t(15;17)(q21;q22) and t(8;21)(q22;q22), detected each in 9% of successful cases, followed by t(9;22)(q34;q11) in 7%, t(4;11)(q21.2;q23.2), t(8;14)(p24.1;q32), +21 each in about 1% (Table 4).
Within group of 23 childhood AML the common cytogenetic abnormalities were t(8;21)(q22;q22) (n = 7), followed by t(15;17)(q21;q22) (n = 3). Within group of 55 childhood ALL the cytogenetic abnormalities were t(9;22)(q34;q11) in 3cases, t(4;11)(q21.2;q23.2) in 2 cases, and t(12:21)(p13;q22), t(1;19)(q23;p13) and del(1p34) in 1 case each (Supplemental Table 2). But normal karyotype was majority in childhood patients with AML and ALL.
Within group of 190 adult AML the common cytogenetic abnormalities were t(15;17)(q21;q22) (n = 25), followed by t(8;21)(q22;q22) (n = 21). Within group of 49 adult ALL the common cytogenetic abnormalities were t(9;22)(q34;q11) (n = 14). Also majority of cytogenetic results was normal karyotype (Supplemental Table 2).
In 228 of 325 patients, there was agreement between the results of RT-PCR and conventional karyotyping in 138 of 325 patients, no abnormality was detected by either method. In 90/325 (28%) patients, a total of 14 fusion transcripts were detected by multiplex RT-PCR, in agreement with the karyotype. In a further of 18 patients (6%), where a specific aberration was demonstrated by both methods, the conventional karyotyping revealed additional structural and/or numerical aberrations. The results of multiplex RT-PCR system and conventional karyotyping did not agree in 97 patients (30%, 97/325). Cytogenetically cryptic translocations found only in the multiplex RT-PCR system were detected in 10% (31/325) patients with a normal karyotype or numerical aberrations by conventional cytogenetic analysis. Detailed data of comparison between multiplex RT-PCR system and conventional cytogenetics are summarized in Table 5. Forty-eight (26%) of 186 patients with a negative result by multiplex RT-PCR analysis had cytogenetic aberrations in the conventional karyotyping. In one child case out of the 186 samples, a t(4;11)(q21;q23) was detected by conventional karyotyping and verified by FISH, while the fusion transcript was not detected by multiplex RT-PCR analysis (Tables 5, 6).
In the current study, 35 types of chromosomal abnormalities were noticed in 325 patients with hematological malignancies. A commercially available multiplex RT-PCR system for screening 28 fusion transcripts detected 20 types among them. When excluding complex and numerical chromosomal abnormalities, 8 types of leukemic fusion genes listed in the commercial multiplex RT-PCR system were not found in our patients (Table 7). Three important cytogenetic abnormalities such as t(8;14)(q24; q32), t(3;3)(q21;q26.2) and i(17)(q10) were also detected in the current study, so these chromosomal abnormalities should be included in a multiplex RT-PCR system for detecting leukemic fusion genes.
Conventional cytogenetic analysis is time-consuming, labor intensive, and has relatively lower sensitivity. In addition, cytogenetics reveals enough chromosomal abnormalities and gives global information about genetic alterations, but not submicroscopic genetic lesions. However, multiplex RT-PCR analysis does not require a large amount of patient specimens, can be performed on resting cells, and is very sensitive in detecting rare abnormal cells including cryptic genetic aberrations. Thus, it should be of great potential benefit to bring the PCR methodology up-front in the diagnosis of acute leukemia. But considering the great number of fusion genes and breakpoint variants presently characterized, more than 50 separate PCR reactions are needed for the screening of a patient with a standard procedure (1). Recently, a commercially available multiplex-RT-PCR system (DNA Technology, Aarhus, Denmark) was introduced to detect 28 common leukemic fusion genes and more than 80 splice variants based on the aforementioned publication (1). Data from Denmark using this system which was applied to specimens from 143 patients with a median age of 63 yr (range 0-85 yr; 132 adults, 11 children) showed that chromosomal rearrangements were detected in only 15% (21/143) of the patients (6). However, the current study revealed a higher detection rate (42%) with the same multiplex RT-PCR system, with 39% (83/213) in AML, 50% in ALL and 50% in MPAL. The spectrum of frequency and distribution of leukemic fusion genes in acute leukemia also differed from previous European data with a wider range of chromosomal rearrangements (6). The incidence of leukemia-specific chromosomal rearrangements in childhood AML disclosed 57% (13/23) of cases, whereas about 40% (24/60) of childhood AML cases had a chromosomal rearrangement using the same multiplex RT-PCR system from Austria (7). The detection rate of leukemic fusion genes in our ALL patients (50%) showed a higher incidence than those from Italian data using the same commercial multiplex RT-PCR system: 53% (29/55) and 47% (23/49) of our childhood and adult ALL cases. However, 39% of 170 ALL cases harbored leukemic fusion genes from the Italian study: 39% and 40% of the childhood and adult ALL cases, respectively (8).
The most frequent chromosomal rearrangements in AML patients in the current study were the t(15;17) abnormality (16%, 35/213). This incidence was higher when compared with data from Caucasian (6.5%-10%), Australian (12%), Japanese (11%) and Singapore-Chinese (11%) (9), but similar to the recent Chinese data (14.3%) (10). This discrepancy may be caused by sample size and possible ethnic differences. The presence of geographic heterogeneity of cytogenetic abnormalities in hematological malignancies has required further investigation and a better understanding of the genetic and environmental factors as etiological factors involved in the development of leukemia. As published elsewhere, the frequency and spectrum of chromosomal rearrangements in ALL were similar between the current study and previous reports (10, 11). The balanced translocation of BCR/ABL fusion gene in all ALL cases was the most prevalent abnormality (20%) in the present study. The incidence of BCR/ABL fusion genes disclosed a similar prevalence in the Southwest Oncology Group data (26%) (12), but it was higher than those from Indian (6%) and Taiwan data (8%) (13, 14). In childhood ALL, the most prevalent chromosomal rearrangement was the TEL/AML1 fusion transcript, followed by E2A/PBX, BCR/ABL and MLL1/AF4. By contrast, in adult ALL, the BCR/ABL fusion transcript was the most frequent, followed by MLL1 rearrangements, TEL/AML1, and SET/CAN.
Discordant results between multiplex RT-PCR and conventional cytogenetic analysis were mainly caused by numerical and submicroscopic abnormalities. A commercially available multiplex RT-PCR system for screening 28 fusion transcripts detected 20 types among 35 types of chromosomal abnormalities found in 325 patients with hematological malignancies. The concordance rate between the two systems in the current study was lower than those of previous studies (6, 7). Due to the location of the breakpoints in the telomeric regions of the chromosomes and submicroscopic deletions that occur in approximately 20% to 30% of such cases, this cryptic translocation is difficult to identify by conventional cytogenetics (7). In one child case of the 325 samples in the current study, a t(4;11)(q21;q23) was detected by cytogenetics and verified by FISH analysis while the fusion transcript was not detected by multiplex RT-PCR analysis.
Of particular interest are cases with 3 important cytogenetic abnormalities such as t(8;14)(q24;q32), t(3;3)(q21;q26.2) and i(17)(q10), which were not covered in the commercial multiplex RT-PCR system. These chromosomal abnormalities are very important and representative cytogenetic aberrations in hematologic malignancies including acute leukemia. The t(8;14)(q24; q32) is a representative cytogenetic abnormality in ALL (Burkitt type) and the t(3;3)(q21;q26.2) is a newly introduced recurrent cytogenetic abnormality in AML from 2008 WHO classification. The i(17)(q10) is most frequently found in blast crisis of Philadelphia-positive CML. The i(17q) as the sole cytogenetic abnormality has also been reported in primary myelofibrosis, hypereosinophilic syndrome, and rare cases of myelodysplastic syndrome (MDS) and myeloproliferative neoplasm (MPN) that evolve into acute non-lymphocytic leukemia. According to several reports, MDS/MPN harboring i(17q) as a sole cytogenetic abnormality is considered as a unique subset with an incidence of 0.4%-1.6% of MDS, or 1% of all myeloid cases, which is characterized by a male predominance, severe anemia, hyposegmented neutrophils, increased micromegakaryocytes and a poor prognosis (15, 16). So these chromosomal abnormalities should be included in panels for leukemic fusion genes suitable for the development of new efficient molecular detection systems adapted to Korean leukemia patients.
In conclusion, the current study demonstrates the spectrum and frequency of chromosomal abnormalities in patients with mainly acute leukemia, which are differed from previous studies. Also, this study may offer important implications in the development of new molecular detection system for screening panel, as well as revisions of the current commercially available multiplex RT-PCR system.
Figures and Tables
Fig. 1
Representative chromosomal abnormalities. (A) Cryptic cytogenetic abnormalities were detected only in the multiplex RT-PCR system, which usually disclosed normal karyotype by conventional cytogenetic analysis. Positive band at 174base pair (arrow) in master PCR step turned out TEL/AML1 gene rearrangement by split out multiplex RT-PCR. (B) Some chromosomal abnormalities such as t(3;3) or t(8;14) should be included in multiplex RT-PCR system, which were not covered in the commercially available multiplex RT-PCR system. Arrows indicated t(3;3)(q21;q26.2). M, molecular weight marker.

Table 2
Frequency of genetic aberrations by age group in patients with hematologic malignancies detected by multiplex RT-PCR system

Table 3
Frequency of genetic aberrations by type of hematological malignancy in all patients detected by multiplex RT-PCR system
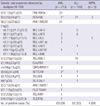
Table 4
Distribution of chromosomal aberrations in participating patients detected by conventional karyotyping including FISH
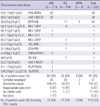
Table 5
Comparison of genetic aberration detected by multiplex RT-PCR system and conventional karyotyping

AUTHOR SUMMARY
Spectra of Chromosomal Aberrations in 325 Leukemia Patients and Implications for the Development of New Molecular Detection Systems
Hyun-Jung Choi, Hye-Ran Kim, Myung-Geun Shin, Hoon Kook, Hyeoung-Joon Kim, Jong-Hee Shin, Soon-Pal Suh and Dong-Wook Ryang
We investigated the chromosomal abnormalities in 325 patients with acute leukemia. The common cytogenetic aberrations in AML patients were PML/PARA > AML1/ETO > MLL1. In ALL patients, the common aberrations were BCR/ABL > TEL/AML1 > MLL1 gene rearrangement, which are differed from previous studies. The results might be useful for the development of new molecular detection system for screening panel as well as diagnosis of acute leukemia.
References
1. Pallisgaard N, Hokland P, Riishøj DC, Pedersen B, Jørgensen P. Multiplex reverse transcription-polymerase chain reaction for simultaneous screening of 29 translocations and chromosomal aberrations in acute leukemia. Blood. 1998. 92:574–588.
2. Bacher U, Kern W, Schnittger S, Hiddemann W, Schoch C, Haferlach T. Further correlations of morphology according to FAB and WHO classification to cytogenetics in de novo acute myeloid leukemia: a study on 2,235 patients. Ann Hematol. 2005. 84:785–791.
3. Betts DR, Ammann RA, Hirt A, Hengartner H, Beck-Popovic M, Kuhne T, Nobile L, Caflisch U, Wacker P, Niggli FK. The prognostic significance of cytogenetic aberrations in childhood acute myeloid leukaemia. A study of the Swiss Paediatric Oncology Group (SPOG). Eur J Haematol. 2007. 78:468–476.
4. Klaus M, Haferlach T, Schnittger S, Kern W, Hiddemann W, Schoch C. Cytogenetic profile in de novo acute myeloid leukemia with FAB subtypes M0, M1, and M2: a study based on 652 cases analyzed with morphology, cytogenetics, and fluorescence in situ hybridization. Cancer Genet Cytogenet. 2004. 155:47–56.
5. Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997. 278:1059–1064.
6. Hutchings Hoffmann M, Wirenfeldt Klausen T, Hasle H, Schmiegelow K, Brondum-Nielsen K, Johnsen HE. Multiplex reverse transcription polymerase chain reaction screening in acute myeloid leukemia detects cytogenetically unrevealed abnormalities of prognostic significance. Haematologica. 2005. 90:984–986.
7. Strehl S, König M, Mann G, Haas OA. Multiplex reverse transcriptase-polymerase chain reaction screening in childhood acute myeloblastic leukemia. Blood. 2001. 97:805–808.
8. Elia L, Mancini M, Moleti L, Meloni G, Buffolino S, Krampera M, De Rossi G, Foà R, Cimino G. A multiplex reverse transcriptase-polymerase chain reaction strategy for the diagnostic molecular screening of chimeric genes: a clinical evaluation on 170 patients with acute lymphoblastic leukemia. Haematologica. 2003. 88:275–279.
9. Nakase K, Bradstock K, Sartor M, Gottlieb D, Byth K, Kita K, Shiku H, Kamada N. Geographic heterogeneity of cellular characteristics of acute myeloid leukemia: a comparative study of Australian and Japanese adult cases. Leukemia. 2000. 14:163–168.
10. Cheng Y, Wang Y, Wang H, Chen Z, Lou J, Xu H, Qian W, Meng H, Lin M, Jin J. Cytogenetic profile of de novo acute myeloid leukemia: a study based on 1432 patients in a single institution of China. Leukemia. 2009. 23:1801–1806.
11. Pui CH. Acute lymphoblastic leukemia in children. Curr Opin Oncol. 2000. 12:3–12.
12. Pullarkat V, Slovak ML, Kopecky KJ, Forman SJ, Appelbaum FR. Impact of cytogenetics on the outcome of adult acute lymphoblastic leukemia: results of Southwest Oncology Group 9400 study. Blood. 2008. 111:2563–2572.
13. Amare P, Gladstone B, Varghese C, Pai S, Advani S. Clinical significance of cytogenetic findings at diagnosis and in remission in childhood and adult acute lymphoblastic leukemia: experience from India. Cancer Genet Cytogenet. 1999. 110:44–53.
14. Chang HH, Lu MY, Jou ST, Lin KH, Tien HF, Lin DT. Cytogenetics in childhood acute lymphoblastic leukemia in Taiwan: a single-institutional experience. Pediatr Hematol Oncol. 2006. 23:495–506.
15. Pinheiro RF, Chauffaille Mde L, Silva MR. Isochromosome 17q in MDS: a marker of a distinct entity. Cancer Genet Cytogenet. 2006. 166:189–190.
16. Kim MH, Hwang HY, Jeong SH, Kim YS, Eo WK, Park JS, Moon YH. Detection of p53 mutant and isochromosome 17q in myelodysplastic syndromes and leukemias. Korean J Clin Pathol. 2000. 20:349–353.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download