Abstract
IgE-dependent activation of mast cells and basophils through the high-affinity IgE receptor (FcεRI) is involved in the pathogenesis of allergen-induced immune responsiveness in atopic diseases like atopic dermatitis (AD). We sought to determine FcεRI gene polymorphisms are associated with AD in Korean patients, and analyzed the relevance of FcεRI gene polymorphisms and serum IgE levels. We conducted a case-control association analysis (175 patients and 56 controls) of Korean subjects. Genotyping was performed using the TaqMan fluorogenic 5' nuclease assay, and serum levels of IgE were measured using a fluorescence enzyme immunoassay. We found that there were no significant relationships between FcεRI and AD, although there were trends towards an association between the 66T>C (rs2251746) polymorphism and total serum IgE levels in the Korean AD patients. In conclusion, while the 66T>C (rs2251746) of the FcεRIα polymorphism may be linked to AD and higher serum IgE levels, polymorphisms in the FcεRIβ gene did not confer susceptibility to AD in our patient sample.
Atopic dermatitis (AD) is a common chronic inflammatory skin disease that, along with asthma and allergic rhinitis, forms the atopic triad (1). It is characterized by xerotic skin with pruritus and chronic inflammation (2). The cause of AD is still unknown, but there is evidence for complex interactions between genetic and environmental factors (3). Genetic factors have an important influence on the risk of developing atopic disease, and several genome-wide searches have provided evidence linking atopy to loci on multiple autosomal chromosomes (4).
High-affinity IgE receptor (FcεRI) is expressed mainly on the surfaces of mast cells, basophils, dendritic cells, Langerhans cells and monocytes (5-7). IgE-dependent activation of mast cells and basophils through the FcεRI is an initial step in the sequence of the atopic reaction (6-10).
FcεRI plays a central role in the induction and maintenance of allergic responses by mast cells and basophils. The β-chain of FcεRI plays a critical role in setting the level of the cellular response to IgE and antigen through its capacity to amplify FcεRI signaling and cell surface expression (11). Several polymorphisms in the gene encoding the β-chain of the FcεRI (FcεRIβ) have been identified (12).
FCεR1A is an important immunity-related gene that encodes the α-chain of FcεRI (13). Two FcεRIα single-nucleotide polymorphisms, -315aC>T (rs2427827) and -66aT>C (rs2251746) appear to be the only common genetic variants localized in a region of the proximal promoter in subjects of European descent (14, 15), and the only two variants present in that region in East Asians have been deemed functional (16-18). In this study, we focused on the two FcεRIα polymorphisms (-315C/T and -66T>C) and the two FcεRIβ polymorphisms (-211C/T and E237G) and performed a genetic association study of susceptibility to atopic dermatitis in Korean subjects.
The present study included 175 patients with AD (80 males and 95 females; mean age 9.18 ± 5.04 yr) and 56 healthy controls (30 males and 26 females; mean age 10.78 ± 6.64 yr). Healthy controls were individuals with serum total IgE levels within the normal range and without AD, allergic asthma, allergic rhinitis, atopic keratoconjunctivitis or other allergic symptoms as determined by a questionnaire. People with present or past atopic disease, IgE-mediated sensitization or a family history of atopy and dermatological diseases were excluded from the control group. All patients fulfilled the Hanifin and Rajka diagnostic criteria for AD (19). The baseline clinical characteristics of the Korean patients with AD are shown in Table 1. As previously described (20, 21), allocation to either the extrinsic (ADe) or intrinsic (ADi) AD phenotype subgroup was performed according to the presence of IgE-mediated sensitization as determined by fluorescence enzyme immunoassay analysis (FEIA) and skin prick test.
The following four FCεR1 gene polymorphisms were typed: FCεR1A-95T/C (rs2251746), FCεR1A-344C>T (rs2427827), MS4A2-109T>C (rs1441586), and MS4A2 E237G (rs569108). All were described previously for a Korean sample (22).
Single nucleotide polymorphism (SNP) genotyping was performed using the TaqMan fluorogenic 5' nuclease assay (ABI, Foster City, CA, USA). Briefly, DNA samples were extracted from whole venous blood (G-DEX™ Genomic DNA Extraction kits; iNtRON Biotechnology, Sungnam, Korea). The extracted DNA was quantified by spectrophotometer, and the quality of the DNA was determined by electrophoresis through an agarose gel and staining with ethidium bromide. The final volume of the polymerase chain reaction (PCR) was 5 µL, containing 10 ng of genomic DNA and 2.5 µL of TaqMan Universal PCR Master Mix, with 0.13 µL of 40 × Assay Mix (Assay ID C___1840470_20, RS2427827-CT4, C___18422260_10, C___2684958_10; ABI). Thermal cycle conditions were as follows: 50℃ for 2 min to activate uracil N-glycosylase and to prevent carry-over contamination, 95℃ for 10 min to activate DNA polymerase followed by 45 cycles of 95℃ for 15 sec and 60℃ for 1 min. All PCR reactions were performed using 384-well plates in a Dual 384-Well GeneAmp PCR System 9700 (ABI), and the endpoint fluorescent readings were performed on an ABI PRISM 7900 HT Sequence Detection System (ABI). Duplicate samples and negative controls were included to ensure genotyping accuracy.
Statistical analyses comparing the allele and genotype distributions were performed using chi-square or Fisher's exact tests. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated according to Woolf's method, using SPSS 10.0 software for Windows (SPSS, Chicago, IL, USA). P < 0.05 was considered statistically significant. Linkage disequilibrium pairwise values were obtained using Haploview software (Broad Institute, Cambridge, MA, USA).
The study protocol was approved by the institutional review board of the Chung-Ang University Hospital Institute Review Board (C2008030/133). Informed consent was obtained from all subjects. All investigations were conducted according to the principles of the Declaration of Helsinki.
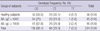
No associations were observed between the -315C>T (rs2427827) polymorphism and the log10-transformed total serum IgE levels in the AD patients in the genotype model (CC vs CT vs TT; 1.97 ± 0.68 vs 2.17 ± 0.61 vs 2.7 ± 0.9, mean ± SD; P = 0.089, unpaired Student's t-test) or the dominant model (1.97 ± 0.68 vs 2.21 ± 0.65; P = 0.078). Recessive and allelic models were not tested because of the very low frequency of -315C>T homozygotes. A comparison of healthy subjects with the entire group of AD patients (n = 175), yielded the following results: CC vs CT vs TT (genotype model) (Table 2), P = 0.36 (chi-square test); CC vs CT + TT (dominant model), P = 0.22 (two-tailed Fisher exact test); CC + CT vs TT (recessive model), P = 0.6 (two-tailed Fisher exact test); C vs T (allelic model), P = 0.16 (two-tailed Fisher exact test) (Table 1). A comparison of the ADe and ADi groups yielded the following results: CC vs CT vs TT (genotype model), P = 0.66 (chi-square test); CC vs CT + TT (dominant model), P = 0.42 (two-tailed Fisher exact test); CC + CT vs TT (recessive model), P = 1.0 (two-tailed Fisher exact test); C vs T (allelic model), P = 0.371 (two-tailed Fisher exact test) (data not shown).
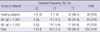
We detected associations between the -66T>C (rs2251746) polymorphism and the log10-transformed total serum IgE levels in the AD patients in a dominant model (TT vs CT + CC; 2.09 ± 0.67 vs 1.82 ± 0.69, mean ± SD; P = 0.026, unpaired Student's t-test). Recessive and genotype models were not tested because of the very low frequency of -66CC homozygotes. A comparison of healthy subjects and the entire group of AD patients yielded the following results: TT vs CT vs CC (genotype model), P = 0.02 (chi-square test); TT vs CT + CC (dominant model), P = 0.07 (two-tailed Fisher exact test); TT + CT vs CC (recessive model), P = 0.25 (two-tailed Fisher exact test); T vs C (allelic model), P = 0.22 (two-tailed Fisher exact test) (Table 3). A comparison of the ADe and ADi groups yielded the following results: TT vs CT vs CC (genotype model), P = 0.8 (chi-square test); TT vs CT + CC (dominant model), P = 0.8 (two-tailed Fisher exact test); T vs C (allelic model), P = 0.64 (two-tailed Fisher exact test).
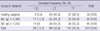
No associations were observed between the -211 C>T (rs1441586) polymorphism and the log10-transformed total serum IgE levels in the AD patients in either the genotype model (TT vs CT vs CC; 2.04 ± 0.68 vs 2.03 ± 0.66 vs 2.06 ± 0.66, mean ± SD; P = 0.989, unpaired Student's t-test) or the dominant model (1.97 ± 0.68 vs 2.21 ± 0.65; P = 0.078). A comparison of healthy subjects and the AD patients yielded the following results: TT vs CT vs CC (genotype model), P = 0.379 (chi-square test); TT vs CT + CC (dominant model), P = 0.706 (two-tailed Fisher exact test); TT + CT vs CC (recessive model), P = 0.243 (two-tailed Fisher exact test); T vs C (allelic model), P = 0.258 (two-tailed Fisher exact test) (Table 4). A comparison of the ADe and ADi groups yielded the following results: TT vs CT vs CC (genotype model), P = 0.871 (chi-square test); TT vs CT + CC (dominant model), P = 0.71 (two-tailed Fisher exact test); TT + CT vs CC (recessive model), P = 0.790 (two-tailed Fisher exact test); T vs C (allelic model), P = 0.781 (two-tailed Fisher exact test).
No associations were observed between the -E237G (rs569108) polymorphism and the log10-transformed total serum IgE levels in the AD patients in either the genotype model (AA vs GA vs GG; 1.97 ± 0.67 vs 2.26 ± 0.71 vs 1.83 ± 0.49, mean ± SD; P = 0.11, unpaired Student's t-test) or the dominant model (1.97 ± 0.67 vs 2.19 ± 0.70; P = 0.121). A comparison of the healthy subjects and the AD patients yielded the following results: AA vs GA vs GG (genotype model), P = 0.753 (chi-square test); AA vs GA + GG (dominant model), P = 0.675 (two-tailed Fisher exact test); AA + GA vs GG (recessive model), P = 1.0 (two-tailed Fisher exact test); A vs G (allelic model), P = 0.477 (two-tailed Fisher exact test) (Table 5). A comparison of the ADe and ADi groups yielded the following results: AA vs GA vs GG (genotype model), P = 0.484 (chi-square test); AA vs GA + GG (dominant model), P = 0.691 (two-tailed Fisher exact test); AA + GA vs GG (recessive model), P = 0.431 (two-tailed Fisher exact test); A vs G (allelic model), P = 1.0 (two-tailed Fisher exact test).
AD is a chronic skin disease that affects more than 15% of children and 2% of adults. Strong connections between genetic factors and AD have been described in the literature. AD is associated with the later development of allergic asthma and rhinoconjunctivitis in approximately 30%-70% of affected individuals (23). AD and other atopic diseases share pathogenic and immunological features, such as elevated total serum IgE and allergen-specific IgE levels. However, the role of IgE in AD still remains unclear (24). Kim et al. (25) indicated that the distribution of the genotype and allele frequencies of the FcεRIβ -109T/C polymorphism is significantly associated with asthma and elevated serum IgE levels. Niwa et al. (26) recently analyzed two promoter polymorphisms of the FcεRIa gene ([FcεRIa], -66T>C [rs2251746] and -315C>T [rs2427827]) in Japanese atopic dermatitis subjects and reported that patients with the -315CT/TT genotype tended to have higher total serum IgE levels, while the proportion of the -315CT/TT genotype or the -315T allele was significantly higher in those with highly elevated total serum IgE. Unlike previous studies, the present study detected no significant differences in allele and genotype frequencies of the four FcεRI polymorphisms between the AD patients and the healthy subjects (Table 2). There were more carriers of the -315C allele among the AD patients than among the healthy subjects, but the difference was not statistically significant. The distributions of all four of the variants did not differ in the ADi patients or in the ADe patients in all tested models. Notably, the distribution of the -66T>C (rs2251746) variant was different between the AD patients and the healthy controls in a genotype model (P = 0.022, chi-square test). However, it did not statistically differ in other tested models, an observation similar to previous findings (16, 26) The distribution of total serum IgE levels in the AD patients was approximated to normal by log10-transformation. There was a trend indicating a relationship between the -66T>C (rs2251746) polymorphism and the log10-transformed total serum IgE levels in the AD patients in a dominant model (TT vs CT + CC; 2.09 ± 0.67 vs 1.82 ± 0.69, mean ± SD; P = 0.026, unpaired Student's t-test). However, neither the number of -66C allele carriers (P = 0.067, two-tailed Fisher exact test) nor the frequency of the -66C allele (P = 0.217, two-tailed Fisher exact test) were significantly different between the AD patients with total serum IgE levels > 1,000 IU/mL and those having total serum IgE concentrations ≤ 1,000 IU/mL (Table 2). No significant relationships were observed between the other polymorphisms (-315C>T, -211C>T and E237G) and the log10-transformed total serum IgE levels in the AD patients. Our findings are not consistent with observations by Potaczek et al. (15), Bae et al. (17) or Hizawa et al. (27), who reported that the FCER1A -315C>T promoter polymorphism and FCER1B genetic variants (-E237G and -109T>C) were associated with total serum IgE concentrations in allergic patients. However, our observation of an association between higher total serum IgE levels and the -66T>C (rs2251746) polymorphism in AD patients concurs with previous reports by Hasegawa et al. (16). They suggested that in a Japanese sample, a statistically significant portion of non-allergic individuals had a heterozygous -66T/C genotype, while most of the allergic individuals had a homozygous -66T/T genotype.
Here we studied the relationship between the FCER1A,B genes and susceptibility to AD in a group of Korean subjects. To our knowledge, this is the first study of the relationship between FCER1A,B and AD in a Korean sample. Our overall data showed no association between FcεRI and AD, although these results may be affected by factors such as genetic background, statistical method, and sample size. In short, according to the findings of the present study, polymorphisms in the FcεRIβ gene do not confer susceptibility to AD in Korean patients, while the 66T>C (rs2251746) variant of the FcεRIα polymorphism may be linked to AD and higher serum IgE levels.
We hypothesize that some discrepancies between our results and the above-mentioned studies may be ascribed to the different origins of the samples analyzed: Polish, Japanese and American. The heterogeneity of American and Polish subjects may be a reason for the differences between our results and those found by Shikanai et al. (14) and Potaczek et al. (15). As a similar example, filaggrin null mutations, which are known for their relevance in atopy, also exhibit population and regional diversity (28-30). Another possible explanation for the differences between our findings and those of other studies may be the small number of patients analyzed in our study, which was 175. If a relationship between a SNP and clinical phenotype was found in a small number of patients, this indicates a very strong association. However, only trends towards an association between the -66T>C (rs2251746) polymorphism and total serum IgE levels in the Korean AD patients were observed. We therefore plan to conduct a more extensive study on two FCER1A loci, the -66T>C (rs2251746) polymorphism that trended towards an association with total serum IgE levels in the Korean AD patients. The -315C>T (rs 2427827) polymorphism was recently analyzed in Japan. This proposed study will include a larger number of patients and will also analyze AD severity, family history, and other clinical aspects.
The present study emphasized the importance of the FCER polymorphism and AD in disease association studies across populations. Further studies of the possible disease-modifying effect of the FCER gene polymorphisms on AD patients are warranted.
Figures and Tables
AUTHOR SUMMARY
FCεRI Gene Promoter Polymorphisms and Total IgE Levels in Susceptibility to Atopic Dermatitis in Korea
Kui Young Park, Mi Kyung Park, Eun Joo Kim, Mi-Kyung Lee and Seong Jun Seo
IgE-dependent activation of mast cells and basophils through the high-affinity IgE receptor (FcεRI) is involved in the pathogenesis of allergen-induced immune responsiveness in atopic diseases like atopic dermatitis. We sought to determine FcεRI gene polymorphisms are associated with AD in Korean patients, and analyzed the relevance of FcεRI gene polymorphisms and serum IgE levels. We conducted a case-control association analysis (175 patients and 56 controls) of South Korean subjects. Genotyping was performed using the TaqMan fluorogenic 5' nuclease assay, and serum levels of IgE were measured using a fluorescence enzyme immunoassay analysis. We found that there were no significant relationships between FcεRI and AD, although there were trends towards an association between the 66T>C (rs2251746) polymorphism and total serum IgE levels in the Korean AD patients. In conclusion, while the 66T>C (rs2251746) of the FcεRIα polymorphism may be linked to AD and higher serum IgE levels, polymorphisms in the FcεRIβ gene did not confer susceptibility to AD in our patient sample.
References
1. Leung DY, Bieber T. Atopic dermatitis. Lancet. 2003. 361:151–160.
2. Kiken DA, Silverberg NB. Atopic dermatitis in children, part 1: epidemiology, clinical features, and complications. Cutis. 2006. 78:241–247.
3. Wüthrich B, Cozzio A, Roll A, Senti G, Kündig T, Schmid-Grendelmeier P. Atopic eczema: genetics or environment? Ann Agric Environ Med. 2007. 14:195–201.
4. Vercelli D. Genetic polymorphism in allergy and asthma. Curr Opin Immunol. 2003. 15:609–613.
5. Saini SS, Richardson JJ, Wofsy C, Lavens-Phillips S, Bochner BS, Macglashan DW Jr. Expression and modulation of FcεRIα and FcεRIβ in human blood basophils. J Allergy Clin Immunol. 2001. 107:832–841.
6. MacGlashan D Jr. IgE and FcεRI regulation. Ann N Y Acad Sci. 2005. 1050:73–88.
7. Sihra BS, Kon OM, Grant JA, Kay AB. Expression of high-affinity IgE receptors (FcεRI) on peripheral blood basophils, monocytes, and eosinophils in atopic and nonatopic subjects: relationship to total serum IgE concentrations. J Allergy Clin Immunol. 1997. 99:699–706.
8. Hulett MD, Brinkworth RI, McKenzie IF, Hogarth PM. Fine structure analysis of interaction of FcεRI with IgE. J Biol Chem. 1999. 274:13345–13352.
9. Wurzburg BA, Jardetzky TS. Structural insights into the interactions between human IgE and its high affinity receptor FcεRI. Mol Immunol. 2002. 38:1063–1072.
10. Metzger H, Chen H, Goldstein B, Haleem-Smith H, Inman JK, Peirce M, Torigoe C, Vonakis B, Wofsy C. A quantitative approach to signal transduction. Immunol Lett. 1999. 68:53–57.
11. Hizawa N, Maeda Y, Konno S, Fukui Y, Takahashi D, Nishimura M. Genetic polymorphisms at FCER1B and PAI-1 and asthma susceptibility. Clin Exp Allergy. 2006. 36:872–876.
12. Nishiyama C, Akizawa Y, Nishiyama N, Tokura T, Kawada H, Mitsuishi K, Hasegawa M, Ito T, Nakano N, Okamoto A, Takagi A, Yagita H, Okumura K, Ogawa H. Polymorphisms in the Fc epsilon RI beta promoter region affecting transcription activity: a possible promoter-dependent mechanism for association between Fc epsilon RI beta and atopy. J Immunol. 2004. 173:6458–6464.
13. Zhang M, Murphy RF, Agrawai DK. Decoding IgE Fc receptors. Immunol Res. 2007. 37:1–16.
14. Shikanai T, Silverman ES, Morse BW, Lilly CM, Inoue H, Drazen JM. Sequence variants in the FcepsilonRI alpha chain gene. J Appl Physiol. 2002. 93:37–41.
15. Potaczek DP, Sanak M, Mastalerz L, Setkowicz M, Kaczor M, Nizankowska E, Szczeklik A. The alpha-chain of high-affinity receptor for IgE (FcepsilonRIalpha) gene polymorphisms and serum IgE levels. Allergy. 2006. 61:1230–1233.
16. Hasegawa M, Nishiyama C, Nishiyama M, Akizawa Y, Mitsuishi K, Ito T, Kawada H, Furukawa S, Ra C, Okumura K, Ogawa H. A novel -66T/C polymorphism in Fc epsilon RI alpha-chain promoter affecting the transcription activity: possible relationship to allergic diseases. J Immunol. 2003. 171:1927–1933.
17. Bae JS, Kim SH, Ye YM, Yoon HJ, Suh CH, Nahm DH, Park HS. Significant association of FcepsilonRIalpha promoter polymorphisms with aspirin-intolerant chronic urticaria. J Allergy Clin Immunol. 2007. 119:449–456.
18. Kanada S, Nakano N, Potaczek DP, Maeda K, Shimokawa N, Niwa Y, Fukai T, Sanak M, Szczeklik A, Yagita H, Okumura K, Ogawa H, Nishiyama C. Two different transcription factors discriminate the -315>CT polymorphism of the Fc epsilon RI alpha gene: binding of Sp1 to -315C and of a high mobility group-related molecule to -315T. J Immunol. 2008. 180:8204–8210.
19. Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol. 1980. 92:44–47.
20. Park JH, Choi YL, Namkung JH, Kim WS, Lee JH, Park HJ, Lee ES, Yang JM. Characteristics of extrinsic vs. intrinsic atopic dermatitis in infancy: correlations with laboratory variables. Br J Dermatol. 2006. 155:778–783.
21. Ott H, Stanzel S, Ocklenburg C, Merk HF, Baron JM, Lehmann S. Total serum immunoglobulin E as a parameter to differentiate between intrinsic and extrinsic atopic dermatitis in children. Acta Derm Venereol. 2009. 89:257–261.
22. Palikhe NS, Kim SH, Cho BY, Ye YM, Hur GY, Park HS. Association of three sets of high-affinity IgE receptor (FcepsilonR1) polymorphisms with aspirin-intolerant asthma. Respir Med. 2008. 102:1132–1139.
23. Williams HC, Wüthrich B. Williams HC, editor. The natural history of atopic dermatitis. Atopic dermatitis. The epidemiology, causes, and prevention of atopic eczema. 2000. Cambridge: Cambridge University Press;41–59.
24. Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol. 2003. 112:6 Suppl. S118–S127.
25. Kim ES, Kim SH, Kim KW, Park HS, Shin ES, Lee JE, Sohn MH, Kim KE. Involvement of FcεR1β gene polymorphisms in susceptibility to atopy in Korean children with asthma. Eur J Pediatr. 2009. 168:1483–1490.
26. Niwa Y, Potaczek DP, Kanada S, Takagi A, Shimokawa N, Ito T, Mitsuishi K, Okubo Y, Tajima M, Hobo A, Ng W, Tsuboi R, Ikeda S, Ogawa H, Okumura K, Nishiyama C. FcepsilonR1alpha gene (FCER1A) promoter polymorphisms and total serum IgE levels in Japanese atopic dermatitis patients. Int J Immunogenet. 2010. 37:139–141.
27. Hizawa N, Yamaguchi E, Jinushi E, Kawakami Y. A common FCER1B gene promoter polymorphism influences total serum IgE levels in a Japanese population. Am J Respir Crit Care Med. 2000. 161:906–909.
28. Ching GK, Hon KL, Ng PC, Leung TF. Filaggrin null mutations in childhood atopic dermatitis among the Chinese. Int J Immunogenet. 2009. 36:251–254.
29. Nomura T, Akiyama M, Sandilands A, Nemoto-Hasebe I, Sakai K, Nagasaki A, Ota M, Hata H, Evans AT, Palmer CN, Shimizu H, McLean WH. Specific filaggrin mutations cause ichthyosis vulgaris and are significantly associated with atopic dermatitis in Japan. J Invest Dermatol. 2008. 128:1436–1441.
30. Barker JN, Palmer CN, Zhao Y, Liao H, Hull PR, Lee SP, Allen MH, Meggitt SJ, Reynolds NJ, Trembath RC, McLean WH. Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J Invest Dermatol. 2007. 127:564–567.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download