Abstract
The size variation of the cytoxin-associated protein (cagA), which is dependent on the 3' repeat region (3'RR) of the cagA gene, is known to play a crucial role in the pathogenesis of Helicobacter pylori infection. The present study evaluated the relationship between the 3'RR variation and the geographic distribution, clinical manifestations, and locations of colonization in the stomach. We evaluated the 3'RR of H. pylori isolates from 78 patients with gastric cancer, peptic ulcer, and non-ulcer dyspepsia from Japan, Hong Kong, India, and the United States and assessed the variations of 3'RR according to the geographical and clinical characteristics. Sixty eight (87.2%) patients had the same 650 bp band without geographical differences. The frequency of polymorphisms in the 3'RR did not differ when compared to the clinical manifestations (P=0.868). The length of 3'RR did not differ by location of colonization. In conclusion, the 3'RR variation of cagA gene is not associated with the geographical and clinical characteristics of the patients studied.
Helicobacter pylori, commonly colonizes the human stomach, and plays a crucial role in the pathogenesis of peptic ulcer disease, gastric cancer and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (1-7). Although many studies have been performed on H. pylori, the pathogenesis of H. pylori infection has not been determined. However, several factors such as the vacuolating cytotoxin (vacA) and the cytotoxin-associated protein (cagA) have been implicated in H. pylori virulence (1-10). The cryptic gene cagA, not present in all H. pylori isolates, is a marker for H. pylori isolates with enhanced interaction with epithelial cells; it encodes a highly immunogenic, variably sized cagA protein (120-140 kDa) (3). The size variation, of the cagA protein, has been correlated with the presence of several repeat sequences located in the 3' region of the repeat reading frame (9). Sequencing of the cagA gene has revealed that the 3' repeat region (3'RR) has a variable fragment length due to internal duplication (3, 6, 11). Although the biological function of 3'RR is not known, 3'RR is thought to escape immunity by generation of either antigenic diversity or immunodominant nonprotective epitopes (3, 6, 10). Prior studies have indicated that some repetitive types of 3'RR are related to gastric histological changes. And recent genetic analysis has revealed that the length of the 3'RR is associated with gastric disease, such as peptic ulcer, gastric atrophy and cancer (12, 13). However, it is not clear whether variations of the 3'RR are responsible for certain clinical outcomes, changes in the gastric environment or whether the variation of the 3'RR on cagA is related to geographic differences. Therefore, the aim of this study was to determine whether the 3'RR variations are related to geographic or clinical manifestations, and whether the variations occur in the same isolates colonizing different locations of the stomach.
We examined 78 patients from four countries; 25 Asian patients from Hong Kong (9 patients with duodenal ulcer [DU], 8 patients with gastric cancer [GC] and 8 patients with non-ulcer dyspepsia [NUD]), 17 white patients from the USA (7 patients with DU and 10 patients with NUD), 8 Indian patients from India (8 patients with NUD), and 28 Asian patients from Japan (16 patients with DU, 6 patients with GC and 6 patients with NUD). Biopsies were taken from the gastric cardia, body and antrum in the patients from the USA and India. Biopsies were obtained from the gastric antrum in the patients from the other countries. The present study was approved by the institutional review board at Kyungpook National University Hospital and informed consent regarding the study and any invasive procedures was obtained from all the patients.
The biopsy specimens were cultured on Trypticase soy agar plates containing 5% sheep's blood (BBL Microbiology Systems, Cockeysville, MD, USA) at 37℃ in ambient air containing 5% CO2 for at least 5 days under microaerobic conditions (Campy-pak systems; BBL, Cockeysville, MD, USA). For growth in the liquid culture, Brucella broth containing 10% FBS supplemented with 50 mM potassium phosphate buffer was inoculated with H. pylori cells harvested from 48 hr Trypticase soy agar/sheep's blood plates and inoculated for 24 hr at 37℃ in a 5% CO2 atmosphere. After 24 hr incubation, the organisms were identified as H. pylori on the basis of Gram stain morphology, colony morphology and positive urease, catalase and oxidase activities.
Chromosomal DNA was prepared from the cells of each isolate after 48 hr of growth on two agar plates; the genomic DNA from the cultures of the isolates was extracted by guanidine thiocyanate (GES, EDTA and sarkosyl), chloroform extraction and isopropanol precipitation. The polymerase chain reaction (PCR) and sequencing was performed with sets of oligonucleotide primers shown in Table 1. The primers were designed based on the known cagA sequences of H. pylori strain 84-183 (Genbank accession number 211714) (Fig. 1). Because diverse isolates were expected in various races from geographic diversity, primers for cag3/4 and cag5/6 were used differently. The cag5/6 primers had a broader range than the cag3/4 among these geographically diverse isolates. The PCR reaction was performed with 10 mM Tris/HCl (pH 8.3), 1.5 mM MgCl2, 200 µM (each) deoxynucleotides, 2 U of Ampli taq polymer, and 1 ng of chromosomal DNA from isolates used as template. The PCR cycling conditions were 35 cycles consisting of 1 min at 95℃, 1 min at 50℃ and 1 min at 72℃. The final cycle included a 7 min extension step to ensure full extension of the PCR product. After purification of the PCR products, using the QlAquick PCR purification kit (Qiagen, Valencia, CA, USA), they were sequenced by an automated sequencer. Primers 5/6 were used to sequence both strands of the DNA fragments. Sequence analysis was performed using the sequencer 3.0 program (Sequencher 3.0, GeneCodes, Ann Arbor, MI, USA). Chromosomal DNA was prepared from bacteria harvested after 48 hr of growth on the Trypticase soy agar. The DNA concentrations were measured by fluorimetry (Dyna Quant 200, Hoefer, San Francisco, CA, USA); 100 ng was used as template for the RAPD-PCR. The primers 1254 (CCGCAGCCAA) and 1281 (AACGCGCAAC) were used, and the results were analyzed by gel electrophoresis as described previously (14).
Seventy eight patients from four countries were examined using two primer sets to amplify the 3'RR of cagA from H. pylori isolates taken from gastic antrum (Table 2). Overall, 68 (87.2%) patients had a 650 bp band, eight patients had bands from 50 to 100 bp or greater, and two had bands 50 bp to 100 bp or shorter. Twenty four out of 25 patients from Hong Kong, and 13 of 17 patients from the USA had the 650 bp band; 8 Indian patients and 23 Japanese patients had the same bands. These results suggest that variation of the 3'RR of cagA gene was not related to geographical differences (P=0.289).
In patients with peptic ulcer disease, 28 out of 32 patients had a 650 bp band, one had a 100 bp shorter band, and three had bands from 50 to 100 bp greater. Among the patients with gastric cancer, 12 of 14 had a 650 bp, and two had a 750 bp band. Twenty eight out of 32 patients with non-ulcer dyspepsia had a 650 bp band, one had a 550 bp band and three had bands from 50 to 100 bp or greater. There was little variation in the frequency (12.5% vs. 14.3%, P=0.868) of polymorphisms. Some of the isolates from the three different clinical diseases showed not only shorter but also larger products. Therefore, clinical differences did not appear to affect the 3'RR of the cagA gene.
Isolates of H. pylori were identified from culture taken from the gastric cardia, body and antrum of the patients from India and the USA (Table 3). Among total 31 isolates from 13 patients, all 19 isolates from 8 Indian patients had a 650 bp band, and the 11 isolates from 5 patients from the USA had a 650 bp bands and one isolate had a 550 bp band. In two isolates from one patient (147 strain), the antral isolates (147 antral strain) had a 550 bp band and the cardia isolates (147 cardia strain) had a 650 bp bands (Fig. 2). To confirm whether the two strains with differences in the 3'RR were the same, RAPD-PCR was performed for fingerprinting. The RAPD-PCR showed an identical profile in the antral and cardia isolates (Fig. 3). These findings indicated that they were clonal variants of one another. The study of single colonies from the antrum and cardia, with the primer cag3/4 set, showed that the three antral isolates had a 550 bp band, whereas the five cardia isolates had a 650 bp band (Fig. 4).
The genetic analysis showed that the cardia isolate was characterized by two repeats of a 15 bp sequence (Rl, EPIYA) and by a 42 bp region (R2, KVNKKKYGVQVASLE) located between the two R1 repeats, and a 147 bp segment (R3, QVAKKVKAKIDRLDQIASGLGDVGQAANFLLKRHDKVDDLSKVGRSVSP) was immediately followed by another R1 sequence (Fig. 5). In R3, downstream of the 3'RR region, we found a 43 bp sequence that was a direct repeat of the same sequence (R4, LKRHDKVDDLSKVG). The 3'RR sequence analysis of the antral isolate showed a 100 bp defect in the distal part of 3'RR compared to the cardia isolate. The deletion was at the distal part of the 3'RR from the inner portion of R4 and R3 to before the start of R4 at the distal of 3'RR.
The CagA gene is a putative marker of virulence; its specific function is unknown (15, 16). PCR has demonstrated size variation of the cagA gene in various H. pylori isolates (3, 6, 11). This is consistent with the size heterogeneity of cagA detected by immunoblot in recent studies (3, 17, 18). The sequence analysis and Southern blotting has shown that the cagA gene contains several repeat sequences in the 3'RR. Differences in this region have been suggested to generate proteins with different sizes and immunogenicity (10, 16, 19, 20). We evaluated whether the variability in the fragment length of the 3'region of cagA was associated with geographical and clinical differences, and whether the variations occur in the same isolates colonizing different locations of the stomach. The results of this study showed about 87% (68/78) of all isolates from four countries had the same PCR product size; the variable sizes of 10 other isolates were scattered evenly without significant differences observed between the Western and Asian isolates. This finding is consistent with prior reports from Western studies (7, 9, 10, 13, 21). Most of the isolates from the three Western and two Asian countries had almost the same size of the 3'RR. Therefore, geographical differences in the 3'RR appear to be very rare.
Although the molecular size appears to be stable, some sequence differences have been identified in Western and Asian isolates without a molecular size change (10). Several studies have reported that the size variation, in the 3'RR of the cagA gene, was associated with specific H. pylori-related gastric disease; the larger size variations were associated with gastric carcinoma and atrophy (9, 10, 22). One African study showed that the length of this region was shorter in isolates from patients with peptic ulcer disease compared to patients with gastritis alone; in addition, the fragment length was longer in isolates from patients with gastric cancer compared to the patients with peptic ulcer or gastritis (21). Another study showed that the longer cagA sizes were found in acid sensitive isolates, which was thought to reflect a strong selective pressure on the isolates that colonize the host to enhance the development of gastritis (23).
However, the results of our study differ from these prior studies. We found that the length of the 3'RR, from almost all isolates from three different clinical diseases was very similar, and that 10 of the isolates from each of the three different diseases had both shorter and longer lengths. These results suggest that the length of the 3'RR was not associated with the disease entities studied, duodenal ulcer, gastric cancer and non-ulcer dyspepsia (20, 24, 25). The 10 isolates showed different molecular sizes, 8 had longer product sizes and 2 had shorter ones. It is likely that the eight isolates with longer cagA had additional internal repeats and the two isolates with shorter cagA likely had deletions in the 3'RR.
In the cagA 3'RR study of multiple isolates from different gastric locations, in the same patients, one of the two shorter isolates was an antral isolate, from the cardia and antral isolates of one patient from the USA. The sequence analysis showed that the antral isolates had about a 100 bp deletion in the 3'RR compared to the same cardia isolate. This explains why the antral isolate was 100 bp-shorter compared to the cardia isolate, and supports our suggestion that the isolates with smaller size PCR products compared to the others, had a deletion in the 3'RR. Our study findings also showed that variation occurs in the same isolates colonizing different locations of the stomach without correlation to the clinical findings and the geographic differences.
The results of this study also showed that the 3'RR from the antral isolate, an area of high acidity, was shorter than the isolates from the cardia. This is consistent with other studies that reported that the length of isolates from peptic ulcer disease with high acidity was shorter than from atrophic gastritis or gastric cancer (9, 10, 22).
In conclusion, the results of present study indicate that the length of the 3'RR is not associated with clinical findings or geographic variations. However, there could be many virulent factors of H. pylori and host factors for various gastrointestinal diseases, and biologic activity of cagA protein should be taken into consideration for further study.
Figures and Tables
Fig. 2
PCR of 3' repeat region of 84-183 and 147 strain's cagA gene by primer cag3/4. 84-183 strains show 50 bp differences and 147 strain from different location of same stomach show 100 bp differences.
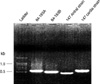
Fig. 3
RAPD-PCR, using primers 1254 and 1281, of antral and cardia isolates from patient 147. Strain 26695 and H2O are controls. The two isolates have the same profiles indicating that they are the same strain.
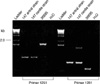
Fig. 4
PCR of 8 isolates from antrum (lanes a-c) and cardia (lanes d-h) of patient 147 using primers cag3 and cag4. All 3 antral isolates showed products shorter than the cardia isolates.

Fig. 5
Primary structure of the cagA 3' repeat region of the isolates from USA patient 147. (A) cardia strain, (B) antral strain. In the genetic analysis, the antral strain showed a 100 bp defect in the cagA 3' repeat region. R3 and downstream of 3' repeat region have a directly repeated same sequence (R4).

References
1. Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998. 28:37–53.

2. Atherton JC. The clinical relevance of strain types of Helicobacter pylori. Gut. 1997. 40:701–703.

3. Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993. 90:5791–5795.

4. Cover TL, Dooley CP, Blaser MJ. Characterization of and human serologic response to proteins in Helicobacter pylori broth culture supernatants with vacuolizing cytotoxin activity. Infect Immun. 1990. 58:603–610.

5. Soltermann A, Koetzer S, Eigenmann F, Komminoth P. Correlation of Helicobacter pylori virulence genotypes vacA and cagA with histological parameters of gastritis and patient's age. Mod Pathol. 2007. 20:878–883.

6. Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993. 61:1799–1809.

7. Umit H, Tezel A, Bukavaz S, Unsal G, Otkun M, Soylu AR, Tucer D, Bilgi S. The relationship between virulence factors of Helicobacter pylori and severity of gastritis in infected patients. Dig Dis Sci. 2009. 54:103–110.

8. Valmaseda Perez T, Gisbert JP, Pajares Garcia JM. Geographic differences and the role of cagA gene in gastroduodenal diseases associated with Helicobacter pylori infection. Rev Esp Enferm Dig. 2001. 93:471–480.
9. Yamaoka Y, El-Zimaity HM, Gutierrez O, Figura N, Kim JG, Kodama T, Kashima K, Graham DY. Relationship between the cagA 3' repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology. 1999. 117:342–349.

10. Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR. Variants of the 3' region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998. 36:2258–2263.
11. Maeda S, Kanai F, Ogura K, Yoshida H, Ikenoue T, Takahashi M, Kawabe T, Shiratori Y, Omata M. High seropositivity of anti-CagA antibody in Helicobacter pylori-infected patients irrelevant to peptic ulcers and normal mucosa in Japan. Dig Dis Sci. 1997. 42:1841–1847.
12. Saruc M, Demir MA, Kucukmetin N, Kandiloglu AR, Akarca US, Yuceyar H. Histological and clinical predictive value of determination of tissue CagA status by PCR in Helicobacter pylori infected patients; results of the large population based study in western Turkey. Hepatogastroenterology. 2002. 49:878–881.
13. Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999. 37:2274–2279.

14. Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992. 20:5137–5142.
15. Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996. 93:14648–14653.

16. Tummuru MK, Sharma SA, Blaser MJ. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol. 1995. 18:867–876.

17. Weel JF, van der Hulst RW, Gerrits Y, Roorda P, Feller M, Dankert J, Tytgat GN, van der Ende A. The interrelationship between cytotoxin-associated gene A, vacuolating cytotoxin, and Helicobacter pylori-related diseases. J Infect Dis. 1996. 173:1171–1175.

18. Xiang Z, Censini S, Bayeli PF, Telford JL, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995. 63:94–98.

19. Cover TL, Blaser MJ. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med. 1996. 41:85–117.
20. Zhou J, Zhang J, Xu C, He L. CagA genotype and variants in Chinese Helicobacter pylori strains and relationship to gastroduodenal diseases. J Med Microbiol. 2004. 53:231–235.

21. Kidd M, Lastovica AJ, Atherton JC, Louw JA. Heterogeneity in the Helicobacter pylori vacA and cagA genes: association with gastroduodenal disease in South Africa? Gut. 1999. 45:499–502.

22. Rudi J, Kolb C, Maiwald M, Kuck D, Sieg A, Galle PR, Stremmel W. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998. 36:944–948.
23. Karita M, Matsumoto S, Kamei T. The size of cagA based on repeat sequence has the responsibility of the location of Helicobacter pylori in the gastric mucus and the degree of gastric mucosal inflammation. Microbiol Immunol. 2003. 47:619–630.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download