Abstract
The hepatitis B virus (HBV) polymerase gene has overlapping reading frames with surface genes, which allows to alter the amino acid codon of the surface genes. In adefovir (ADV) treated chronic hepatitis B patients carrying rtA181T/rtA181V mutations, overlap with surface gene mutations such as sW172stop/sL173F has been reported. However, the clinical consequences of such surface mutations have not been determined. The aim of this study was to determine the surface gene sequence in ADV-resistant patients carrying the A181T/V mutation and to describe the clinical significance. Of the 22 patients included in this study, 13 were ADV-resistant with rtA181T/V mutations (polymerase mutation group, Group P) and nine were antiviral treatment-naïve (control group, Group C). The Pre-S1 gene mutation, V60A, was detected in 11 patients (Group P=8, Group C=3). A start codon mutation in the Pre-S2 gene was found in five patients (Group P=3, Group C=2). An S gene mutation, sA184V, was found in nine patients, all of whom were in group P. Although sW172stop and sL173F mutations were detected, reduced HBsAg titer was not observed. Further study of these mutations and their clinical implications are needed.
Oral antiviral agents are currently available and are the main option for the treatment of patients with chronic hepatitis B. In a large proportion of patients, antiviral therapy can slow down the disease progression by hepatitis B virus (HBV) DNA suppression and hepatitis 'e' antigen (HBeAg) seroconversion. However, the potential emergence of drug resistant mutations remains a caveat of oral antivirals; mutations increase as the treatment duration extends (1, 2). In addition, mutations to one drug may lead to resistance to other drugs. Patients with lamivudine (LMV)-resistant rtM204I/V (±rtL180M) mutations in the catalytic tyrosine-methionine-aspartate-aspartate (YMDD) motif are less responsive to entecavir (ETV) (1, 2). Among adefovir (ADV) treated patients, rtA181T or rtA181V mutations have been associated with a 2-6 fold decrease in sensitivity to LMV (2).
Mutation of the HBV polymerase can alter the amino acid codon of the surface genes and vice versa, because the polymerase gene, the main target of antiviral drug resistant mutations, has overlapping reading frames with the surface region (1, 3). In LMV treated patients, the P120A mutation in the S region results in hepatitis B surface antigen (HBsAg) detection failure (4). Lee et al. (5) reported overlapping surface gene mutations of HBV in an LMV-treated patient who finally lost HBsAg. In this patient, they detected deletions of nucleotides 23 to 55 (amino acids 12 to 22) in the Pre-S2 region, and amino acid substitutions at I126S, T131N, M133T, and S136Y in the 'a' determinant region of HBsAg.
Overlap of surface gene mutations, such as sW172stop in patients carrying rtA181T, and sL173F in patients carrying rtA181V, have been reported in chronic hepatitis B patients receiving ADV treatment. However, the clinical consequences of these changes have not been determined (1, 2). The goal of this study was to evaluate the surface gene sequence in ADV-resistant patients carrying A181T/V mutations and to determine the associated clinical significance.
Twenty-two patients with chronic hepatitis B were included in this study. Nine patients had not been previously treated with antiviral agents and thus formed the control group (nucleos[-t]ide naïve patients, Group C). The remaining thirteen patients were ADV-resistant, having an rtA181T mutation, rtA181V mutation, or both mutations (ADV polymerase mutation group, Group P). These patients developed ADV resistance while undergoing ADV monotherapy as rescue therapy for LMV-resistant HBV (rtM204I/V±rtL180M). The ADV-resistant mutations were detected using restriction fragment mass polymorphism (RFMP) method as previously described (6). Serum was collected from the patients every two to three months during treatment and was kept at -20℃ until the mutation analyses were performed. The serum collected at the time of ADV-resistant rtA181T/V mutation detection was used for sequencing analysis. Informed consent was obtained from each patient and the experimental protocol was approved by the Korea University Guro Hospital Human Research Committee.
At the time of serum collection, we conducted routine full blood examinations and liver function tests, including tests for aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, albumin, prothrombin time, blood urea nitrogen (BUN), and creatinine. HBsAg and antibody to the HBs antigen were measured using the commercially available radioimmunoassay kits (Abbot Laboratories, North Chicago, IL, USA). The HBsAg titer was measured using the chemiluminescent microparticle immunoassay (CMIA) ARCHITECT® HBsAg assay (Abbot Laboratories). HBeAg and antibody to the HBe antigen were measured using commercially available radioimmunoassay kits (Shin Jin Medics Inc., Goyang, Korea). HBV-DNA was quantified using the Hybrid Capture II assay (Digene Diagnostics Inc., Beltsville, MD, USA) or the VERSANT® HBV DNA 3.0 assay (Siemens Healthcare Diagnostics Inc., Tarrytown, NY, USA) and data showed 'pg/mL' were converted to 'copies/mL' (1 pg=283,000 copies).
Hepatitis B viral DNA was extracted from 200 µL of serum using the QIAamp® MinElute® Virus Spin kit (Qiagen Inc., Vanencia, CA, USA) according to the manufacturer's instructions. The S coding region of HBV was amplified using HBS-1S (the forward primer) and HBS-1AS (the backward primer) as previously described (5). The amplification conditions consisted of 94℃ for 3 min, 35 cycles of 94℃ for 30 sec, and 55℃ for 30 sec. This was followed by a final primer extension at 72℃ for 1 min using the Taq PCR Core Kit (Qiagen Inc.). The polymerase chain reaction (PCR) amplified HBV DNA was purified using the QIAquick® PCR Purification Kit (Qiagen Inc.) according to the manufacturer's instructions.
The primers used for direct sequencing were HBS-1S, HBS-1AS, HBS-2S, and HBS-3S as previously described (5). Sequencing reactions were performed in an MJ Research PTC-225 Peltier Thermal Cycler using an ABI PRISM BigDye™ Terminator Cycle Sequencing Kit with AmpliTaq DNA polymerase (Applied Biosystems Inc., Foster, CA, USA) according to the manufacturer's instructions. Single-pass sequencing was performed on each template using the aforementioned primers. The fluorescent-labeled fragments were purified from the unincorporated terminators with an ethanol precipitation protocol. The samples were resuspended in distilled water and subjected to electrophoresis in an ABI PRISM 3730XL Analyzer (Applied Biosystems Inc.).
The nucleotide sequences were compared with those of the HBV genotype C subtype 'adr' registered at the National Center for Biotechnology Information (NCBI, nucleotide LOCUS AF286594). The genotype of the obtained HBV DNA sequences was determined by a web-based genotyping tool for viral sequences at the NCBI (7).
We conducted the Fisher's exact tests for categorical variables and Mann-Whitney's U-tests for continuous variables to compare patients with the rtA181T/V mutation to those without the rtA181T/V mutation. Values are expressed as median (range). A P value <0.05 was considered statistically significant.
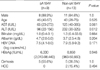
Demographic and biochemical data are summarized in Table 1. Among the 13 patients in Group P, the majority were males (n=10). All patients in Group C were males. Biochemical data are represented as median (range) (median of Group P vs. Group C, P value). Group P was significantly older than Group C, 45 (28-75) (47 vs. 39, P=0.025), had higher levels of albumin 4.2 (2.5-5) (4.2 vs. 3.6, P>0.05). Group P also had lower levels of AST 100.5 (23-385) (65 vs. 193, P=0.001), ALT 155 (22-385) (78 vs. 262, P<0.001), and total bilirubin 1.1 (0.4-33.6) (1.0 vs. 1.3, P>0.05). Ten of the 22 patients had liver cirrhosis (LC) (Group P=7, Group C=3). Thirteen of the 22 patients were HBeAg positive (Group P=7, Group C=6). Two cases of hepatocellular carcinoma (HCC) were included (Group P=1, Group C=1).
All patients in our study were infected with HBV genotype C. Twenty-one of the 22 patients had the 'adr' subtype and one patient had the 'ayr' subtype. For the Pre-S1, Pre-S2, and S genes, the incidence of mutations in Group P and Group C were 3 vs. 3 (P>0.05), 1 vs. 3 (P>0.05), and 4 vs. 2 (P>0.05), respectively. The median number of point mutations in the entire region was 8 (Group P=8, Group C=10, P>0.05). Most cases had mutations in the S gene except for one case in Group C. One patient in Group P did not have a mutation in the Pre-S1 gene and seven patients (Group P=5, Group C=2) had no mutations in the Pre-S2 gene (Table 2).
We conducted serum HBsAg titers to evaluate the clinical consequences of the surface gene mutation. The median HBsAg titer was 7,590 IU/mL (Group P=5,950, Group C=15,090, P>0.05). There was no significant difference, regardless of whether they harbored the rtA181T/V mutation (Table 2).
In the Pre-S1 region, 12 patients (six cases in each group) had nucleotide deletions at position '21' (amino acid [a.a.] 7) and 18 patients (nine cases in each group) had nucleotide deletions at position '30' (a.a. 10). Nucleotide deletions at position '59' (a.a. 20) were detected in 5 of 13 patients in Group P and 2 of 9 patients in Group C. An H51Q mutation was detected in two patients (one case in each group). Eight of 13 patients in Group P and 3 of 9 patients in Group C had a V60A mutation. G2A (n=1), G73D (n=1), L74I (n=2), A91T (n=1), and A95T (n=1) mutations were detected in Group P. In Group C, A81T (n=1), T86A (n=2), and A95V (n=1) mutations were detected. Three cases of I84T mutations were found in Group C, compared to only one in Group P (Fig. 1).
A start codon mutation was identified in five patients (Group P=3, Group C=2) in the Pre-S2 gene. All five patients with this mutation had cirrhosis, and two of these patients also had HCC (Group P=1, Group C=1). V32A, I45T, and T49I mutations were detected in the patients with HCC, both groups equally. An F46S mutation was observed in Group P (n=2); a Q2K mutation in Groups P (n=1) and C (n=1); an A11T mutation in Groups P (n=2) and C (n=1); and an F22L mutation in Groups P (n=2) and C (n=2).
In Group C, there was one case of each of the following mutations: N4K, T7A, H9Q, R16P, R16G, V17P, V17A, R18S, G19S, L20Q, P54Q, nucleotide 418-423 deletion (a.a. 21-22), 419 deletion (a.a. 21), and 425 deletion (a.a. 23). In Group P, mutations I42T, P54L, and nucleotide 406 deletion (a.a. 17) were detected (Fig. 2).
Among the patients in Group P, sW172stop (n=5) and sL173F (n=5) mutations were detected. In the S gene, an A184V mutation was found in 9 of 13 patients in Group P. However, it was not detected in Group C (P=0.002). Other mutations that we detected in the S gene include S3N (Group P=4, Group C=2), I68T (Group P=3, Group C=1), I126T (Group P=4, Group C=1), and L213I (Group P=6, Group C=1). These mutations were most frequent in Group P.
In Group C, there was one case of each of the following mutations: G10stop, L15V, I28V, W36L, F41S, T47A, Q51L, N59H, C69stop, L87V, F93L, V96G, L97P, G102C, T131P, Y200F, and C221Y. Similarly, in Group P, there was one case of each of the following mutations: F41C, A45V, P49L, Q54L, P62L, I92T, L98V, T140S, A157V, W182stop, and S204R. An L21S mutation was detected in Groups P (n=2) and C (n=2), and R24Q and P203R mutations were found in Groups P (n=1) and C (n=1) (Figs. 3, 4).
Among these mutations, only the W172stop/L173F (P<0.001) and A184V (P=0.002) mutation were significantly different between the groups (data not shown).
We also compared patients both with and without the W172stop/L173F mutation (Table 3). AST (P=0.035) and ALT (P=0.009) levels were significantly lower in patients with W172stop/L173F mutation. However, there was no significant difference when the same comparison was done in Group P alone.
We compared patients both with and without the sA184V mutation (Table 4). Only the ALT level was significantly lower in patients with the sA184V mutation (P=0.012). However, there was no significant difference when the same comparison was done in Group P alone.
HBV is an enveloped virus and contains a partially double-stranded DNA genome of approximately 3,200 base pairs (1, 3, 8). The major envelope protein of HBsAg consists of 226 amino acids and is coded by the S gene (9). The polymerase gene overlaps all six other genes, including the three envelope genes, Pre-S1, Pre-S2, and S, which encode for the large (by the Pre-S1/S2/S gene), middle (by the Pre-S2/S gene), and small (the S gene) envelope proteins, respectively (1, 10). Due to the overlap between the polymerase and surface genes, the selection of a drug-resistant HBV polymerase gene mutant may have important clinical and diagnostic implications (1, 3).
Deletions and mutations of overlapping surface genes have been reported in LMV treated patients who loose HBsAg (3). In addition to surface gene deletions and mutations, deletions of nucleotides in the Pre-S2 region and amino acid substitutions in the 'a' determinant region of HBsAg have been reported (5). The LMV-resistant polymerase mutation, rtV173L (sE164D), has been reported to reduce the antigenicity of HBsAg (11).
Drug resistant mutations to ADV have been reported mainly in the HBV polymerase domain D rtN236T or the domain B rtA181V/T (1, 12). Whereas a domain D rtN236T mutation does not overlap with the envelope gene, a mutation at rtA181T can result in a stop mutation in the envelope region of the S gene (sW172stop). In addition, an ADV-resistant mutation at rtA181V results in a concomitant change at sL-173F (1). Therefore, we selected LMV-resistant chronic hepatitis B patients who had developed ADV-resistant A181T/V mutations during ADV rescue therapy. We sought to detect other mutations caused by the ADV-resistant mutation and its clinical implications. Patients who had not received antiviral treatment were selected for the control group.
In the present study, biochemical data showed that AST, ALT and total bilirubin levels were higher in Group C. And it can be considered that Group C has relatively worse biochemical level. One limitation should be noted: we used indirect markers or surrogates of liver function in the biochemical tests and fibrosis, proliferation, apoptosis, and other pathogenic processes were not measured. The patients should be followed for a long period of time and the clinical impact of the rtA181T/V mutation should be evaluated.
The results of our study showed that treatment induced or naturally occurring mutations of the Pre-S1, Pre-S2, and surface antigen regions were detected in most of the patients studied. In Group P, although statistically insignificant, Pre-S1 region nucleotide deletions at position '59', and mutations V60A and L74I were the primary changes detected. Five patients carrying Pre-S2 start codon mutations had either cirrhosis or HCC. In addition, V32A, I45T, and T49I mutations at Pre-S2 were detected in the HCC patients only. An F46S mutation in the Pre-S2 region was detected only in patients with the ADV-resistant rtA181T/V mutation. These results are consistent with a number of previous reports indicating that Pre-S mutations are associated with advanced liver disease and active viral replication (13-19). In Group C, a number of point mutations were detected. A similar pattern of mutations was also detected in Group P. There were no specific mutations confined to the Pre-S2 region in Group C. Nucleotide 21 deletions were frequently observed in both Groups P and C, and nucleotide 30 deletions were observed in all but four cases. We are uncertain as to what caused the sequence changes. It is difficult to suggest that the sequence changes were caused by resistant mutations because these deletions were more common in Group C. The development of Pre-S deletion mutants depends on HBV genotypes (20). Recently, one report showed that Pre-S deletions were more common in Korean HBV genotype strains and correlated with genotype C and advanced liver disease (21). The mutations in the Pre-S region, particularly the deletions, may affect the ratio between the small and large envelope proteins, resulting in endoplasmic reticulum stress associated with the aggravation of liver disease (21, 22). These patients should be observed closely.
The S gene mutation, A184V, was detected in the ADV-resistant rtA181T/V polymerase mutation group alone. This mutation site overlaps with the rt192 region in the polymerase gene. However, the A184V mutation did not result in amino acid changes in the polymerase region. There is one published report that the A184V mutation was related to reduced or negative HBsAg signal (23). In our study, however, changes in HBsAg titer were not observed. When compared with patients who have not A184V mutation including Group C, ALT level alone was significantly lower in patients with A184V mutation. But it is difficult to conclude that it is caused by A184V mutation, because this difference was no longer significant when the analysis was done in pure Group P and patient's number was not enough. The clinical implications of this remain to be determined. Of all mutations within the 'a' determinant region of HBsAg, the most common antigenic determinant mutations are in amino acids 124 to 147 (24). Substitutions within the 'a' loop can create a putative glycosylation site in mutant HBVs that result in undetectable HBsAg antigenicity (25). In our study, an I126T mutation was detected in 4 of 13 ADV mutants and one of nine control cases. One case of a T131P mutation in Group C and one case of a T140S mutation in Group P were also detected. Some investigators have reported that s126 threonine is a wild type variant and nucleotide substitution of threonine has no specific effect (26-29). To determine the clinical significance of I126T, T131P, and T140S mutations, further studies are required. It is possible that these amino acid substitutions were caused by a remaining LMV-resistant mutation, though a rtM204I/V mutation was not detected in sequencing analysis.
Although surface gene mutations were common in Group P, serum HBsAg titers were not significantly different from those of Group C. Despite the fact that we did not measure in vitro hepatitis B surface antigen production, the impact of coexisting wild type virus, viral replication, fitness, packaging, and antigenic targets for immunoassay should be considered. Full-length HBV genomes should be cloned and the S gene should be sequenced and analyzed. It should be noted that, in this study, patients who were nucleoside treatment-naïve controls had a variety of Pre-S gene mutations as well as surface gene mutations.
We detected sW172stop and sL173F mutations in 10 of the 13 patients with ADV-resistant rtA181T/V polymerase mutations. A large percentage of the cases with rtA181T mutations developed sW172stop mutations. In cases with ADV treated LMV-resistant mutations, the rtA181T mutation was reported at the ADV treatment baseline with low HBV DNA titers (30).
In conclusion, the results of this study showed that treatment-induced or naturally occurring mutations of the Pre-S1, Pre-S2 and surface antigen regions were frequent. The sA184V mutation was detected in the A181T/V polymerase mutation group alone. Although sW172stop and sL173F mutations were detected in the polymerase mutation group, a reduction in HBsAg titer was not found. Further study is needed to determine the clinical implications of viral replication, topographic alteration, fitness, envelope gene production, and interaction with wild type HBV DNA.
Figures and Tables
Fig. 1
Sequence changes in the Pre-S1 regions. Twelve patients (six cases in each group) and 18 patients (nine cases in each group) had nucleotide deletions at positions '21' and '30'. Nucleotide deletions at position '59' were detected in 5 of 13 patients in Group P and in 2 of 9 patients in Group C. An H51Q mutation was detected in two patients (one case in each group). A V60A mutation was detected in 8 of 13 patients in Group P and 3 of 9 patients in Group C. L74I mutations in Group P (n=2) and T86A mutations in Group C (n=2) were detected. Three cases of an I84T mutation were found in Group C and one case in Group P. G2A, G73D, A91T, and A95T mutations were detected in Group P. A81T and A95V mutations were detected in Group C.

Fig. 2
Sequence changes in the Pre-S2 regions. Five start codon mutations were identified in the Pre-S2 region. V32A, I45T and T49I mutations were detected in HCC cases. Q2K (Group P=1, Group C=1), A11T (Group P=2, Group C=1), F22L (Group P=2, Group C=2), and F46S (Group P=2) mutations were observed. In Group C, there were following mutations: N4K, T7A, H9Q, R16P, R16G, V17P, V17A, R18S, G19S, L20Q, P54Q, nucleotide '418-423' deletion, '419' deletion, and '425' deletion. In Group P, one case each of I42T, P54L mutation and nucleotide 406 deletion were detected.

Fig. 3
Sequence changes in the S regions up to the front of 'a' determinant. An S3N mutation in Groups P (n=4) and C (n=2), and an I68T mutation in Groups P (n=3) and C (n=1) were detected. G10stop, L15V, I28V, W36L, F41S, T47A, Q51L, N59H, C69-stop, L87V, F93L, V96G, L97P, and G102C mutations were detected in Group C (one case each). F41C, A45V, P49L, Q54L, P62L, I92T, and L98V mutations were identified in Group P (one case each). An L21S mutation in Groups P (n=2) and C (n=2) and an R24Q mutation in Groups P (n=1) and C (n=1) were detected.

Fig. 4
Sequence changes in the S regions from 'a' determinant. In 10 of 13 patients in Group P, sW172stop (n=5) and sL173F (n=5) mutations were detected. A184V mutations were found in 9 of 13 patients in Group P. I126T mutations in groups P (n=4) and C (n=1) and L213I mutations in Groups P (n=6) and C (n=1) were detected. T131P, Y200F, and C221Y mutations were detected in Group C (one case each). T140S, A157V, W182stop, and S204R mutations were identified in Group P (one case each). One P203R mutation was detected in each group.

References
1. Bartholomeusz A, Locarnini S. Hepatitis B virus mutations associated with antiviral therapy. J Med Virol. 2006. 78:Suppl 1. S52–S55.

2. Locarnini S, Mason WS. Cellular and virological mechanisms of HBV drug resistance. J Hepatol. 2006. 44:422–431.

3. Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J Clin Virol. 2002. 25:97–106.

4. Hsu CW, Yeh CT, Chang ML, Liaw YF. Identification of a hepatitis B virus S gene mutant in lamivudine-treated patients experiencing HBsAg seroclearance. Gastroenterology. 2007. 132:543–550.

5. Lee SY, Choi MS, Lee D, Lee JH, Koh KC, Paik SW, Yoo BC. Overlapping gene mutations of hepatitis B virus in a chronic hepatitis B patient with hepatitis B surface antigen loss during lamivudine therapy. J Korean Med Sci. 2005. 20:433–437.

6. Yeon JE, Yoo W, Hong SP, Chang YJ, Yu SK, Kim JH, Seo YS, Chung HJ, Moon MS, Kim SO, Byun KS, Lee CH. Resistance to adefovir dipivoxil in lamivudine resistant chronic hepatitis B patients treated with adefovir dipivoxil. Gut. 2006. 55:1488–1495.

7. Rozanov M, Plikat U, Chappey C, Kochergin A, Tatusova T. A web-based genotyping resource for viral sequences. Nucleic Acids Res. 2004. 32:W654–W659.

8. Summers J, O'Connell A, Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc Natl Acad Sci USA. 1975. 72:4597–4601.

9. Michel ML, Tiollais P. Structure and expression of the hepatitis B virus genome. Hepatology. 1987. 7:1 Suppl. 61S–63S.

11. Torresi J, Earnest-Silveira L, Deliyannis G, Edgtton K, Zhuang H, Locarnini SA, Fyfe J, Sozzi T, Jackson DC. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology. 2002. 293:305–313.

12. Angus P, Vaughan R, Xiong S, Yang H, Delaney W, Gibbs C, Brosgart C, Colledge D, Edwards R, Ayres A, Bartholomeusz A, Locarnini S. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology. 2003. 125:292–297.

13. Choi MS, Kim DY, Lee DH, Lee JH, Koh KC, Paik SW, Rhee JC, Yoo BC. Clinical significance of pre-S mutations in patients with genotype C hepatitis B virus infection. J Viral Hepat. 2007. 14:161–168.

14. Chen CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, Lu SN, Changchien CS. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology. 2007. 133:1466–1474.

15. Raimondo G, Costantino L, Caccamo G, Pollicino T, Squadrito G, Cacciola I, Brancatelli S. Non-sequencing molecular approaches to identify preS2-defective hepatitis B virus variants proved to be associated with severe liver diseases. J Hepatol. 2004. 40:515–519.

16. Preikschat P, Gunther S, Reinhold S, Will H, Budde K, Neumayer HH, Kruger DH, Meisel H. Complex HBV populations with mutations in core promoter, C gene, and pre-S region are associated with development of cirrhosis in long-term renal transplant recipients. Hepatology. 2002. 35:466–477.

17. Fan YF, Lu CC, Chen WC, Yao WJ, Wang HC, Chang TT, Lei HY, Shiau AL, Su IJ. Prevalence and significance of hepatitis B virus (HBV) pre-S mutants in serum and liver at different replicative stages of chronic HBV infection. Hepatology. 2001. 33:277–286.

18. Pollicino T, Zanetti AR, Cacciola I, Petit MA, Smedile A, Campo S, Sagliocca L, Pasquali M, Tanzi E, Longo G, Raimondo G. Pre-S2 defective hepatitis B virus infection in patients with fulminant hepatitis. Hepatology. 1997. 26:495–499.

19. Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006. 130:1153–1168.

20. Sugauchi F, Ohno T, Orito E, Sakugawa H, Ichida T, Komatsu M, Kuramitsu T, Ueda R, Miyakawa Y, Mizokami M. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol. 2003. 70:537–544.

21. Mun HS, Lee SA, Jee Y, Kim H, Park JH, Song BC, Yoon JH, Kim YJ, Lee HS, Hyun JW, Hwang ES, Kook YH, Kim BJ. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008. 80:1189–1194.

22. Caselmann WH, Meyer M, Kekule AS, Lauer U, Hofschneider PH, Koshy R. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc Natl Acad Sci USA. 1990. 87:2970–2974.

23. Olinger CM, Weber B, Otegbayo JA, Ammerlaan W, van der Taelem-Brule N, Muller CP. Hepatitis B virus genotype E surface antigen detection with different immunoassays and diagnostic impact of mutations in the preS/S gene. Med Microbiol Immunol. 2007. 196:247–252.

24. Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990. 336:325–329.

25. Koyanagi T, Nakamuta M, Sakai H, Sugimoto R, Enjoji M, Koto K, Iwamoto H, Kumazawa T, Mukaide M, Nawata H. Analysis of HBs antigen negative variant of hepatitis B virus: unique substitutions, Glu129 to Asp and Gly145 to Ala in the surface antigen gene. Med Sci Monit. 2000. 6:1165–1169.
26. Ohnuma H, Machida A, Okamoto H, Tsuda F, Sakamoto M, Tanaka T, Miyakawa Y, Mayumi M. Allelic subtypic determinants of hepatitis B surface antigen (i and t) that are distinct from d/y or w/r. J Virol. 1993. 67:927–932.

27. Yamamoto K, Horikita M, Tsuda F, Itoh K, Akahane Y, Yotsumoto S, Okamoto H, Miyakawa Y, Mayumi M. Naturally occurring escape mutants of hepatitis B virus with various mutations in the S gene in carriers seropositive for antibody to hepatitis B surface antigen. J Virol. 1994. 68:2671–2676.

28. Okamoto H, Yano K, Nozaki Y, Matsui A, Miyazaki H, Yamamoto K, Tsuda F, Machida A, Mishiro S. Mutations within the S gene of hepatitis B virus transmitted from mothers to babies immunized with hepatitis B immune globulin and vaccine. Pediatr Res. 1992. 32:264–268.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download