Abstract
Community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) is an increasingly common worldwide and colonizing S. aureus strains may serve as the causative pathogen for overt clinical infections. This study was performed to determine whether the pathogenic CA-MRSA isolate in clinical infections was genetically related to the MRSA isolates in community carriers. We prospectively collected a total of 42 CA-MRSA isolates (23 clinical infection isolates and 19 colonization isolates) in a local region of Korea. Antimicrobial susceptibility tests, staphylococcal toxin assays, SCCmec typing, multilocus sequence typing (MLST), and spa (staphylococcal protein A) typing were performed with all isolates. Thirty-four (81%) of 42 CA-MRSA isolates belonged to sequence type (ST) 72 in the MLST analysis. The distribution of STs did not differ significantly between colonization and clinical infection isolates (89.5% [17/19] vs. 73.9% [17/23], P=0.26). Among the ST72-MRSA isolates, spa type t664 (18, 52.9%) and t324 (8, 23.5%) were common in both groups. This study demonstrates that the community-associated MRSA strains from patients with clinical infections are closely related to the strains found in carriers from one local community.
Since the 1980s, methicillin-resistant Staphylococcus aureus(MRSA) infections have become a serious clinical problem among hospitalized patients worldwide. In Korea, the frequency of methicillin resistance among S. aureus isolates from Korean tertiary-care hospitals was reported to account for more than 70% of all cases in 2003 (1). Since the first case of community-associated methicillin-resistant S. aureus (CA-MRSA) infection was reported in the United States in 1980 (2), recent studies have reported that CA-MRSA infections appeared to be on the increase in both adults and children in various regions and countries (3-8). CA-MRSA isolates in the United States and Australia have been reported to possess a novel staphylococcal cassette chromosome mec (SCCmec) type, type IV, and frequently contain a gene coding for Panton-Valentine leukocidin (PVL) (9).
Colonizing S. aureus strains may serve as the causative pathogen for overt clinical infections and may spread to other patients in a hospital (10). Nasal S. aureus carriage has been identified as a risk factor for the pathogenesis of community-acquired and nosocomial infection (11). However, previous studies have not examined the genetic relationship between S. aureus strains from patients with clinical infections and strains from colonized individuals in the community.
Recently, molecular typing techniques such as pulsedfield gel electrophoresis (PFGE), multilocus sequence typing (MLST), and spa (staphylococcal protein A) typing have been used to document the local and global epidemiology of MRSA isolates (12-15). PFGE has been used the most widely, but the results obtained from different laboratories often can not be compared (16). Therefore DNA sequence typing, including MLST and spa typing, has become more popular (17). In this study, we evaluated the genetic relationship between CA-MRSA strains from carriers and those from patients with staphylococcal infection using the MLST and spa typing.
We had prospectively collected all MRSA isolates from 2004 through 2006 in a tertiary care hospital in Gyeongsang Province, Korea. All isolates were obtained from patients who were admitted to the hospital or from individuals who visited the Health Promotion Center of this hospital. MRSA isolates were considered to be community-associated if the isolate was obtained from a patient who had no risk factors for health care-associated infection and the sample was collected within 72 hr of admission. Established risk factors for health care-associated infection included a history of hospitalization, surgery, dialysis, or residence in a long-term care facility within one year prior to the MRSA_culture date, or the presence of a permanent indwelling catheter or medical device (e.g. tracheostomy tube, gastrostomy tube, or Foley catheter) at the time of the culture collection (6). Clinical infection was assumed if a patient with an MRSA isolate presented with clinical manifestations of staphylococcal infection in the bloodstream, central nervous system, bone, joints, lungs, pleural fluid, muscle, skin, or soft tissue. Colonization was assumed if the MRSA was isolated from a patient lacking clinical signs and symptoms of a staphylococcal infection or from an individual who was screened for MRSA carriage from the anterior nares or throat in the Health Promotion Center.
Antimicrobial susceptibility testing was performed at 35℃ on Mueller-Hinton agar by the disk diffusion method, and the results were interpreted using Clinical and Laboratory Standards Institute (CLSI) breakpoint criteria (18). All strains were tested for the presence of the mecA gene by a multiplex PCR (determined using the two PCR methods described below).
Genomic DNAs of S. aureus were prepared using the Qiagen DNA Minikit (Qiagen, Hilden, Germany), according to the manufacturer's recommendations. PCR amplification for the staphylococcal enterotoxin genes (sea, seb, sec, sed, and see), toxic shock syndrome toxin gene (tst), exfoliative toxin genes (eta and etb), and PVL gene (pvl) was performed as previously reported (19). SCCmec typing was performed using the multiplex PCR method of Oliveira and de Lencastre (20). This SCCmec typing method can amplify nine different genes in one reaction, and subtypes are determined by the presence or absence of these genes.
MLST was performed as described previously (12). PCR fragments of the seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) were obtained from chromosomal DNA and were directly sequenced. The allelic profiles of the S. aureusstrains were defined on the basis of their MLST type. Each unique allelic profile was designated a sequence type (ST), which was determined using the database at the MLST website (http://saureus.mlst.net/).
Spa typing was performed as previously described (21). DNA sequences were obtained with an ABI 377 Sequencer (Applied Biosystems, Foster City, CA, USA). spa types were determined using the database at the Ridom SpaServer website (http://spa.ridom.de/spatypes.shtml).
A total of 42 non-duplicated CA-MRSA isolates were collected from 2004 through 2006: 12 isolates from 2004, 12 isolates from 2005, and 18 isolates from 2006 (Fig. 1). Of these 42 isolates, 19 were collected from the clinical specimens of patients without clinical signs and symptoms of staphylococcal infection (n=10: 6 from urine and 4 from sputum) and from the MRSA screening culture samples from the throat or anterior nares (n=9: 5 from throat swab and 4 from nasal swab). Twenty-three isolates were taken from patients with clinical MRSA infections: skin and soft-tissue infection (n=8), bacteremia (n=6), urinary tract infection (n=2), pneumonia (n=2), and other clinical infections (n=5).
The majority of the 42 CA-MRSA isolates did not have any toxin genes. The sea was detected in 2 clinical infection isolates with ST239 clone and 2 isolates (1 clinical infection and 1 colonization isolate) with ST1 clone. The tst was detected in 1 clinical infection isolate with ST5 clone. The pvl was not detected in any of the isolates.
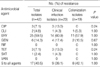
The MLST analysis of the 42 CA-MRSA isolates revealed five ST types, including ST72, ST1, ST5, ST239 and ST89. The most prevalent ST was ST72 (34/42, 81%). The incidence of ST72 in clinical infection isolates was similar to that in colonization isolates (73.9% [17/23] vs. 89.5% [17/19]). There were seven spa types in our ST72-MRSA isolates: t664, UJGGMDMGM (n=18, 52.9%); t324, UJGGMDMGGM (n=8, 23.5%); t901, UJGMDMGGM (n=2, 5.9%); t2473, UJGFGM (n=1, 2.9%); undetermined type (UT), UJGGM (n=3, 8.8%); UT, UJGGMDMGGMM (n=1, 2.9%); and UT, UJGGDMGM (n=1, 2.9%). These ST72-MRSA isolates showed deletion-insertion events in spa, which generated seven spa types (Table 1). ST5 and ST239 clones were detected in 4 isolates from only clinical infection group; the spa type of two ST5-MRSA isolates was t002, TJMBMDMGMK, and that of the two ST239-MRSA isolates was t37, WGKAOMQ. Three isolates (2 from blood and 1 from urine) which were obtained from neonates who were less than 30 days of age had ST1-MRSA-IV, spa type t286, UJEBKBPE.
The SCCmec types identified in the CA-MRSA isolates included: type IVa (n=30, 71.4%), IV (n=8, 19.5%), III (n=2, 4.8%), II (n=1, 2.4%), and IIa (n=1, 2.4%). There were four different SCCmec types (IVa, II, III, and IV) in the pathogenic isolates and type IVa predominated (65.2%). Of 19 colonization isolates, there were three SCCmec types (types IVa, IV, and IIa) and the most common type was also type IVa (78.9%). ST72 MRSA strains, which predominated in all CA-MRSA isolates in this study, were SCCmec type IVa or IV, and the majority of them were SCCmec type IVa (29 of 34, 85.3%).
The overall rates of resistance to erythromycin, gentamicin, ciprofloxacin, tetracycline, clindamycin, and trimethoprim-sulfamethoxazole among the CA-MRSA isolates were 59.5%, 14.3%, 9.5%, 7.1%, 4.8%, and 2.4%, respectively. None of the isolates were resistant to rifampin or vancomycin. The majority (85.7%) of the CA-MRSA isolates from both groups were susceptible to all antimicrobial agents we tested, or were only resistant to erythromycin. The resistance rates for each antimicrobial agent did not differ significantly between groups (Table 2).
Molecular methods have demonstrated a close relationship between those CA-MRSA strains causing clinical infection and strains from colonized individuals who represent the prevalent CA-MRSA clone in one Korean community.
There are distinct genetic backgrounds of CA-MRSA associated with each geographic origin and the molecular types of CA-MRSA strains do not correspond to those of HA-MRSA within each continent (5). MLST and spa type analyses have indicated that there are CA-MRSA strains with ST30 in Australian and South American isolates, ST80 in European and Middle Eastern isolates, ST59 in Taiwanese isolates, and ST1 ST8 and ST59 in the USA isolates (5, 22).
Recently, a report on CA-MRSA infections in Korea showed that the ST72-MRSA-IVa, spa types t664 and t324, were found in the CA-MRSA isolates from children and adults who visited an outpatient clinic in one local region of Korea with diverse staphylococcal infections, even though those clones were not dominant in that report (23). Another study in Korea showed that ST72-MRSA-IVa clones were most common in the MRSA strains isolated from seven community-based or tertiary hospitals (24). The current study has revealed that ST72-MRSA-IVa, spa types t664 and t324 clones form a large majority of the CA-MRSA isolates, and they are found in various clinical isolates regardless of age, sex, isolation site, or whether the isolate was accompanied by the clinical manifestations of MRSA infection or not.
ST72 strains already have been found in a few HA-MRSA isolates from one tertiary care hospital in Korea (25). The ST72-HA-MRSA strains had a genetic background that was identical in terms of molecular typing to the ST72-CA-MRSA strains from the isolates collected in the outpatient clinic (25). A Swedish report recently noted that MRSA isolates with ST72-MRSA-IVa, spa types t664 and t324, were found in screening culture samples of young children from Korea adopted by Swedish families; all adoptees had a history of hospitalization in their native country before arriving in Sweden (26). Whether the ST72-MRSA strains originated in a community or hospital setting, they are now widely distributed in the community in Korea; all ST72-MRSA strains from previous studies and our study were collected in the community.
In 2006, it was reported that ST1-MRSA-IVa, spa type t286, was detected in MRSA isolates from neonates between 1 and 13 days of age, who were transferred from 2 primary obstetrics clinics to a tertiary care hospital in Seoul, Korea (27). ST1-MRSA isolates detected in our study were also obtained from neonates and were spa type t286 clones, even though their SCCmec type (type IV) was different from that of the MRSA isolates detected in the previous study. These findings suggest that ST1-MRSA, spa type t286, might be distributed in the community in Korea, and primary obstetrics clinics may be a reservoir for this MRSA clone.
ST5 and ST239 isolates, which are known to be the dominant genotype of HA-MRSA isolates in Korea, were detected in 4 clinical infection isolates in our study. The patients with these isolates had no risk factors for health care-associated infection in their medical records. We suggest that these ST5 and ST239-MRSA clones may have originated in a hospital reservoir and then spread in the community.
The pvl represents a stable marker of CA-MRSA strains, which is a possible virulence factor associated with necrotic lesions of the skin and subcutaneous tissue (e.g. furuncles) and also with community-acquired severe necrotic pneumonia (9, 28). However, none of the CA-MRSA isolates harbored pvl in our study. This observation is consistent with previous Korean studies (23, 24).
There are several limitations to the current study. First, this investigation was performed at a single tertiary health care center. Therefore, the findings may not be generalized to other regions of Korea. Nevertheless, our data shows a close genetic relationship between CA-MRSA isolates causing clinical infection and the colonized isolates in a local community. Second, our subjects were patients hospitalized in a tertiary care hospital. Thus, generalization of these findings to other populations, especially to patients treated at outpatient clinics in the community, should be done with caution. Third, we analyzed only a relatively small number of CA-MRSA isolates. This might be caused by the relative low incidence rate of CA-MRSA infection in Korea, contrary to our expectation. A small data set may introduce sampling bias, so the findings of this study should not be generalized to the entire population of Korea until additional data are available.
In conclusion, our study shows that CA-MRSA strains from patients with clinical infection are genotypically similar to those from colonized individuals and ST72-MRSA-IVa, spatypes t664 and t324, which are different from the HA-MRSA clones previously reported in Korea, were the prevalent clones among the CA-MRSA isolates collected in one region of Korea.
Figures and Tables
Fig. 1
Enrollment of community-acquired methicillin-resistant Staphylococcus aureus isolates.
*This is the percentage of MRSA isolates among all S. aureus isolates.
HPC, Health promotion center; ES, Elementary school.
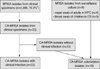
Table 1
Genotypic characteristics between clinical infection and colonization isolates of CA-MRSA

*ST determined based on the MLST website (http://saureus.mlst.net); †Nomenclature according to Shopsin et al. (21); ‡Nomenclature according to Harmsen et al. (13).
ST, sequence type; UT, undetermined type.
References
1. Kim HB, Park WB, Lee KD, Choi YJ, Park SW, Oh MD, Kim EC, Choe KW. Nationwide surveillance for Staphylococcus aureus with reduced susceptibility to vancomycin in Korea. J Clin Microbiol. 2003. 41:2279–2281.
2. Community-acquired methicillin-resistant Staphylococcus aureus infections--Michigan. MMWR Morb Mortal Wkly Rep. 1981. 30:185–187.
3. Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998. 279:593–598.
4. Salmenlinna S, Lyytikainen O, Vuopio-Varkila J. Community-acquired methicillin-resistant Staphylococcus aureus, Finland. Emerg Infect Dis. 2002. 8:602–607.
5. Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003. 9:978–984.
6. Fridkin SK, Hageman JC, Morrison M, Sanza LT, Como-Sabetti K, Jernigan JA, Harriman K, Harrison LH, Lynfield R, Farley MM. Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005. 352:1436–1444.
7. Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis. 2005. 5:275–286.
8. Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006. 44:108–118.
9. Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999. 29:1128–1132.
10. von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001. 344:11–16.
11. Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997. 10:505–520.
12. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000. 38:1008–1015.
13. Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003. 41:5442–5448.
14. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003. 41:5113–5120.
15. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005. 43:5026–5033.
16. Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003. 41:1574–1585.
17. Aires-de-Sousa M, Boye K, de Lencastre H, Deplano A, Enright MC, Etienne J, Friedrich A, Harmsen D, Holmes A, Huijsdens XW, Kearns AM, Mellmann A, Meugnier H, Rasheed JK, Spalburg E, Strommenger B, Struelens MJ, Tenover FC, Thomas J, Vogel U, Westh H, Xu J, Witte W. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J Clin Microbiol. 2006. 44:619–621.

18. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 17th informational supplement. 2007. Wayne, PA: CLSI.
19. Peacock SJ, Moore CE, Justice A, Kantzanou M, Story L, Mackie K, O'Neill G, Day NP. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun. 2002. 70:4987–4996.
20. Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002. 46:2155–2161.
21. Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999. 37:3556–3563.
22. Wang CC, Lo WT, Chu ML, Siu LK. Epidemiological typing of community-acquired methicillin-resistant Staphylococcus aureus isolates from children in Taiwan. Clin Infect Dis. 2004. 39:481–487.
23. Ma SH, Lee YS, Lee SH, Kim HK, Jin JS, Shin EK, Lee JC. Methicillin-resistant Staphylococcus aureus clones with distinct clinical and microbiological features in a Korean community. J Med Microbiol. 2007. 56:866–868.
24. Kim ES, Song JS, Lee HJ, Choe PG, Park KH, Cho JH, Park WB, Kim SH, Bang JH, Kim DM, Park KU, Shin S, Lee MS, Choi HJ, Kim NJ, Kim EC, Oh MD, Kim HB, Choe KW. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J Antimicrob Chemother. 2007. 60:1108–1114.
25. Cha HY, Moon DC, Choi CH, Oh JY, Jeong YS, Lee YC, Seol SY, Cho DT, Chang HH, Kim SW, Lee JC. Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean hospital. J Clin Microbiol. 2005. 43:3610–3614.
26. Gustafsson EB, Ringberg H, Johansson PJ. MRSA in children from foreign countries adopted to Swedish families. Acta Paediatr. 2007. 96:105–108.

27. Ko KS, Park S, Peck KR, Shin EJ, Oh WS, Lee NY, Song JH. Molecular characterization of methicillin-resistant Staphylococcus aureus spread by neonates transferred from primary obstetrics clinics to a tertiary care hospital in Korea. Infect Control Hosp Epidemiol. 2006. 27:593–597.
28. Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, Bes M, Vandenesch F, Piémont Y, Brousse N, Floret D, Etienne J. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002. 359:753–759.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download