Abstract
Authors had prepared the high-quality sectioned images of a cadaver head. For the delineation of each cerebral gyrus, three-dimensional model of the same brain was required. The purpose of this study was to develop the segmentation protocol of cerebral gyri by referring to the three-dimensional model on the personal computer. From the 114 sectioned images (intervals, 1 mm), a cerebral hemisphere was outlined. On MRIcro software, sectioned images including only the cerebral hemisphere were volume reconstructed. The volume model was rotated to capture the lateral, medial, superior, and inferior views of the cerebral hemisphere. On these four views, areas of 33 cerebral gyri were painted with colors. Derived from the painted views, the cerebral gyri in sectioned images were identified and outlined on the Photoshop to prepare segmented images. The segmented images were used for production of volume and surface models of the selected gyri. The segmentation method developed in this research is expected to be applied to other types of images, such as MRIs. Our results of the sectioned and segmented images of the cadaver brain, acquired in the present study, are hopefully utilized for medical learning tools of neuroanatomy.
Perhaps more than tens of thousands magnetic resonance images (MRIs) of brain are being examined and interpreted daily in all parts of the world. Usually in the axial MRIs, neurologists, neurosurgeons, and neuroradiologists are responsible for identifying each cerebral lobe and gyrus for localization of diseases. The lateral sulcus, central sulcus, and some other primary sulci together with neighboring structures are used for the landmarks of localization (1, 2). To get more orientation, the axial MRIs are supplemented with the coronal and sagittal images of the same brain. However, the stereoscopic shape of the convoluted gyri, that are not entirely surrounded by sulci, cannot be readily figured out in the sectional planes. Moreover, individual variations of the gyri make it difficult even to experienced medical experts. Only three-dimensional (3D) model of the same cerebral hemisphere enables final confirmation of the gyri. Fortunately, the 3D volume model is built automatically from the brain MRIs and rotated at free angles on the workstation, included in the MRI scanner.
We made sectioned images of a cadaver head. The sectioned images had extremely small intervals and pixel size (both 0.1 mm) with real human brain color (48 bit color). So the images, the source of state-of-the-art 3D models, were expected to contribute to the neuroanatomy teaching, neurosurgical simulation, and so on. Additionally, basic brain structures were outlined in advance, involving cerebrum, cerebellum, brainstem, etc. (3). Our next goal was to segment the cerebral gyri for functional approach to the cerebrum. Like the MRIs, the 3D model of sectional images was necessary for the full detection of the gyri. But unlike the MRIs, sectioned images could not be volume-reconstructed automatically. So as to build the 3D volume model on the personal computer, we considered to use easily available software, MRIcro (www.mricro.com, version 1.4) (3, 4). The aim of this study is to establish and share the segmentation method of cerebral gyri by referring to the 3D model of same subject on the personal computer.
The sectioned images were made of a cadaver head. We selected a Korean male cadaver who was 67-yr-old, 1,620 mm height, and 45 kg weight. Although he suffered from myasthenia gravis, his head structures with brain were normal. The cadaver head was serially sectioned at 0.1 mm intervals using precise cryomacrotome and the photographed using high quality Canon EOS 5D digital camera to create sectioned images (pixel size, 0.1 mm; color depth, 48 bits color; file format, tag image file format [TIFF] files) (3).
Sectioned images containing only cerebrum were prepared by the previous segmented images. The original number of the sectioned images (TIFF) of head was 2,341 (intervals, 0.1 mm). Segmentation was performed at 1 mm interval to produce 234 segmented images (file format, photoshop document [PSD] files) (3). Among 234 couples of corresponding sectioned and segmented images, we excluded images above or below the cerebrum, so that 114 couples remained. In the images (resolution, 4,368×2,912), excessive margins beyond the cerebrum were cropped to make resolution 2,071×2,064 (Fig. 1A). Using the segmented images of the cerebrum (Fig. 1B), the sectioned images showing just the cerebrum were obtained on Adobe Photoshop CS3 version (Photoshop) (Fig. 1C).
The sectioned images of the cerebrum were processed on Photoshop for the next step of volume reconstruction. The color depth of the images was decreased from 48 bit color to 8 bit gray in order to adapt to MRIcro software. For the same reason, the resolution (2,071×2,064) of the sectioned images (pixel size, 0.1 mm) was reduced to 207×206, which resulted in pixel size of 1 mm, which was identical to the images intervals of 1 mm. All sectioned images of the cerebrum were simultaneously split along the longitudinal cerebral fissure. Consequently, two sets of sectioned images were produced: a set containing the left cerebral hemisphere (resolution, 103×206) and another set containing the right one (resolution, 104×206). The sectioned images of the left hemisphere were printed out on paper for drawing gyri later (Fig. 2E).
The volume model of the cerebral hemisphere was built on MRIcro: Sectioned images were opened by using 'Conversion wizard: convert all files in a folder...', then the volume modeling was automatically carried out to build a volume model (resolution, 103×206×114; voxel size, 1 mm; file format, IMG) (3). By rotating the volume model, four basic views (lateral, medial, superior, inferior) of the cerebral hemisphere were captured and saved as TIFF files (Fig. 2A-D).
All cerebral gyri were determined and painted in the four captured views. On the MRIcro, the volume model was rotated at arbitrary angles to decide the boundaries of all cerebral gyri. The rotation could be done in a real time by mouse dragging thanks to the low resolution of volume model. Territories of the identified gyri were painted on the lateral, medial, superior, and inferior views of the volume model (5, 6). On Photoshop, semitransparent painting was executed with different colors until the entire cerebral cortex was full of colors (Fig. 2A-D). The identified and colored gyri, which counted 33, were classified according to the cerebral surfaces and lobes (Table 1) (7).
Based on the painted views, each gyrus was identified on the printed paper of sectioned images. On the painted lateral and medial views, horizontal grid with 1 mm gaps was overlapped either really or virtually. The grid numbers (1-114) from superior to inferior were same as the serial numbers of the sectioned images with 1 mm intervals. Therefore, in case that grid 100 is overlapped (Fig. 2A, B), gyri in the sectioned image 100 could be recognized without difficulty (Fig. 2E, F). In addition, the painted superior and inferior views were helpful to disclosure of the gyri, located in the superior and inferior ends of cerebral hemisphere, respectively (Fig. 2C, D). On the printed paper, the identified gyri were drawn and annotated by a pen. The boundary of each gyrus included not only cerebral cortex but also cerebral medulla, which was the core of the gyrus (Fig. 2E) (6, 8).
Referring to the drawn papers, the cerebral gyri were delineated in the high-resolution sectioned images (0.1 mm pixel size) at 1 mm intervals. The delineation was performed on the Photoshop in the same manner as our previous paper (3, 9). In the sectioned images, external borders of the gyri were graphically distinguished from the surrounding subarachnoid space, thus they could be drawn semiautomatically; so were some internal borders of the gyri. However, most internal borders were not graphically distinguished, thus drawing was done manually by one of the authors. The outlines of each gyrus were placed on the PSD files' layers, named with the official term of gyrus (Table 1), to achieve the segmented images. After erasing the background sectioned images, the outlines were filled with the colors (Fig. 2F); at this time, the same colors were used as used for painting the gyri in the four captured views (Fig. 2A-D). The segmented images were then converted from PSD files to TIFF files without layers.
Both the sectioned images and segmented images were stacked to acquire coronal and sagittal planes, which were examined for verification of segmentation (Fig. 3). When incorrect segmented images were found, they were revised.
Two segmented images of the left and right cerebral hemispheres were reunited to finalize the segmented images of the entire cerebrum (Fig. 1D). It was processed with 114 couples of images simultaneously using the Photoshop script. The united segmented images were corresponding to the original sectioned images of the head (Fig. 1A). The segmented images of the cerebral gyri, prepared in this research, could be further combined with previous segmented images of other head structures (3).
It took 30 days to produce the 114 segmented images (intervals, 1 mm) of 33 cerebral gyri (Table 1): three days were required for identification of gyri on volume model and for painting on the four captured views (Fig. 2A-D), three days for pen drawing on the printed papers (Fig. 2E), 20 days for delineation on the sectioned images (Fig. 2F), and four days for certification of the computer delineation (Fig. 3B, D). The 30 days were unreasonably long in clinics, but the duration seemed acceptable for acquiring meaningful data in research field.
The segmented images had two file forms, PSD and TIFF. The PSD files included the original sectioned images and outlines on the layers. So the PSD files were helpful for the better automatic production of intended 2D and 3D images (9). The TIFF files, namely color-filled images, were easily distributable because of its much smaller file size. The TIFF files, accompanied by the sectioned images (TIFF files), were able to produce any wanted 2D and 3D images. We decided to release the PSD or TIFF files on each user's demand. The distributed segmented images would be accompanied by four captured views (TIFF files), on which the gyri were painted with the same color as the color-filled images. The four captured views and Table 1 would give users a good orientation of the segmented images of cerebral gyri (Fig. 2).
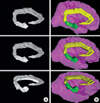
The segmented images were employed for the manufacture of volume model. By using the segmented images (PSD files), sectioned images including selected gyri were produced on Photoshop, then volume model of the gyri was built on MRIcro (Fig. 4A) (3). With programming in computer language, volume model with high resolution and real brain color would be made of the segmented images and original sectioned images.
The segmented images independent of the sectioned images were applied to the surface model producing on Autodesk Maya version 2009 and supplementary software. From the segmented images (PSD files), outlines of each gyrus were stacked. Subsequently gaps between the outlines were filled with polygons, that was the surface reconstruction, to create surface model of the gyrus (10-12). The surface models were satisfactory (Fig. 4B) and corresponding to the volume model of the same gyri (Fig. 4A). It means that the segmented images in this investigation were qualified.
In the neuroimaging field, the segmentation of each cerebral gyrus contributes to neurosurgical simulation, neuroscience research, or other achievements. Current issue in this paper is how to segment the gyrus easily and objectively in the personal computer environment.
For the segmentation, it is required to utilize both two-dimensional (2D) images and 3D image. 3D image of cerebrum is good to categorize every gyrus (Fig. 2A-D); but stereoscopic boundary of the gyrus including the cerebral cortex and subcortical white matter cannot be practically drawn on the 3D image. The 2D images, that are the sectional planes, are good for the precise drawing (Fig. 2E, F); but identification of any given gyrus is extremely difficult on the 2D images. Therefore, both 2D and 3D images of the same cerebrum are desirable to be displayed together.
On the personal computer, our proposed method is efficient in visualizing both 2D and 3D images. MRIcro, freely downloadable, has the useful function to promptly reconstruct the 3D images from the serial 2D images (3). Even if the displayed 3D image is not qualified as the final result, the 3D image is sufficient for the segmentation itself (Fig. 2A-D).
Our proposed method is hopefully welcomed by researchers, who would like to use the personal computer separately from the MRI machine. For instance, researchers could transfer brain MRIs of patients to their laboratories and segment the cerebral gyri for the various kinds of simulations (e.g. stereoscopic surgery or radiotherapy). Researchers could also be interested in non-clinical images, such as the sectioned images from Visible Human Project or the histological serial images from confocal microscopy. Our proposed method on the personal computer is likely to promote convenient segmentation in diverse sorts of images.
In the near future, we expect an advanced program, where simultaneous modification of both 2D and 3D images is done. It means that segmentation on the 2D images automatically reflects modification of the 3D image, and the reverse reflection is possible too.
The segmented images of the cerebral gyri, prepared in this study, might be utilized by many investigators for the following reasons. First, the segmented images deal with all cerebral gyri (Table 1) as well as other structures inside and outside the brain (3). The segmented images can yield 3D surface models of brain gyri and adjacent structures, which are the source of the interactive simulator of neurosurgery (Fig. 4) (11, 12).
Second, the segmented images can be expanded on the basis of the initial sectioned images. Possibly, other brain structures are additionally segmented on the sectioned images according to the different intentions of various investigators. The intervals (1 mm) of the segmented images could be made identical with those (0.1 mm) of the original sectioned images. This trial, enabled by interpolation of the outlines from neighboring segmented images, would cause enhanced 3D images for neuroanatomy (12).
Third, the segmented images are accompanied by the corresponding sectioned images. Two image sets are capable of generating volume models of brain components, which should be realistic enough.
Fourth, the segmented images are also compared with the 7 Tesla MRIs of the same cadaver, scanned before serial sectioning (3). Therefore, it is possible to build a medical learning tool, in which the segmented images as well as the sectioned images help interpretation of the brain MRIs (Fig. 5).
Fifth, the segmented images (both PSD, TIFF), sectioned images, and 7 Tesla MRIs are available free of charge for non-commercial applications. Once approval is obtained for using this data from our group, users all over the world will be provided with the data either on- or off-line.
In conclusion, we developed segmentation method of cerebral gyri with the help of the 3D models on the personal computer. It is hoped that this technique is utilized for the quick and precise segmentation on other images, including brain MRIs. We would like to distribute the segmented images of cerebral gyri together with the corresponding sectioned images to every wanted user.
Figures and Tables
Fig. 1
Sectioned and segmented images generated in sequence. In the original sectioned images of head (A row), only cerebrum is segmented (B row) to acquire the sectioned images of the cerebrum (C row). Each gyrus of the cerebrum is then segmented (D row).
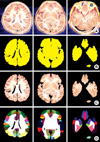
Fig. 2
Gyri segmentation referring to the volume model of the cerebrum. Lateral (A), medial (B), superior (C), and inferior (D) views of the volume model of the left cerebral hemisphere, on which entire cerebral gyri are painted with different colors; printed paper of a sectioned image, on which the cerebral gyri are delineated and annotated by pen (E) to elaborate segmented image of the gyri (F).
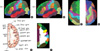
Fig. 3
Coronal, sagittal planes of the sectioned images and segmented images. Coronal sectioned image (A), coronal segmented image (B), sagittal sectioned image (C), and sagittal segmented image (D) are produced for verification of segmentation.

Fig. 4
Volume/surface models of the cingulate and parahippocampal gyri. Volume model of the gyri built on MRIcro (A column) and colored surface model of the gyri and cerebral hemisphere reconstructed on Maya (B column) are slightly rotated.
References
1. Baron JC, Chételat G, Desgranges B, Perchey G, Landeau B, de la Sayette V, Eustache F. In vivo mapping of gray matter loss with voxel-based morphometry in mild Alzheimer's disease. Neuroimage. 2001. 14:298–309.

2. Teipel SJ, Alexander GE, Schapiro MB, Möller HJ, Rapoport SI, Hampel H. Age-related cortical grey matter reductions in non-demented Down's syndrome adults determined by MRI with voxel-based morphometry. Brain. 2004. 127:811–824.

3. Park JS, Chung MS, Shin DS, Har DH, Cho ZH, Kim YB, Han JY, Chi JG. Sectioned images of the cadaver head including the brain and correspondences with ultrahigh field 7.0 T MRIs. Proc IEEE. 2009. 97:1988–1996.
4. Park JS, Chung MS, Park HS, Shin DS, Har DH, Cho ZH, Kim YB, Han JY, Chi JG. A proposal of new reference system for the standard axial, sagittal, coronal planes of brain based on the serially-sectioned images. J Korean Med Sci. 2010. 25:135–141.

5. Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to Cerebral Imaging. 1988. Stuttgart: Thieme.
6. Cho ZH. 7.0 Tesla MRI Brain Atlas: In Vivo Atlas With Cryomacrotome Correlation. 2009. New York, Dordrecht, Heidelberg, London: Springer.
7. Federative Committee on Anatomical Terminology. Terminologia Anatomica: International Anatomical Terminology. 1998. Stuttgart, New York: Thieme.
8. Kiernan JA. Barr's the Human Nervous System: An Anatomical Viewpoint. 2009. 9th Ed. Philadelphia, Baltimore, New York, London, Buenos Aires, Hong Kong, Sydney, Tokyo: Lippincott Williams & Wilkins;211–218.
9. Park JS, Chung MS, Hwang SB, Lee YS, Har DH. Technical report on semiautomatic segmentation using the Adobe Photoshop. J Digit Imaging. 2005. 18:333–343.

10. Park JS, Shin DS, Chung MS, Hwang SB, Chung J. Technique of semiautomatic surface reconstruction of the Visible Korean Human data by using commercial software. Clin Anat. 2007. 20:871–879.
11. Shin DS, Chung MS, Lee JW, Park JS, Chung J, Lee SB, Lee SH. Advanced surface reconstruction technique to build detailed surface models of liver and neighboring structures from the Visible Korean Human. J Korean Med Sci. 2009. 24:375–383.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download