Abstract
We investigated whether the detection of prostate specific membrane antigen (PSMA) in blood preoperatively has predictive value for biochemical recurrence (BCR) after radical prostatectomy in patients with prostate cancer. All 134 patients scheduled to receive radical prostatectomy for prostate cancer were prospectively enrolled. The authors used nested reverse transcriptase-polymerase chain reaction (RT-PCR) assay to detect PSMA mRNA-bearing cells in peripheral blood, and analyzed the ability of PSMA mRNA positivity to predict BCR after surgery. PSMA-mRNA was detected in 24 (17.9%) patients by RT-PCR. Over a median follow-up of 20 months (range, 3 to 46 months), BCR developed in 15 patients (11.2%) and median time to BCR was 7 months (range, 3 to 25 months). Kaplan-Meier analysis revealed a significant difference between those positive or negative for PSMA in terms of recurrence-free actuarial probability (log rank P=0.0039). Multivariate analysis showed that positivity for PSMA mRNA (HR: 3.697, 95% CI 1.285-10.634, P=0.015) and a biopsy Gleason score of ≥7 (HR: 4.500, 95% CI 1.419-14.274, P=0.011) were independent preoperative predictors of BCR. The presence of PSMA mRNA in peripheral blood can be used to predict BCR after radical prostatectomy.
Radical prostatectomy is considered a standard treatment for patients with localized prostate cancer. However, some patients experience recurrence after surgery, and this is known to be associated with prostate-specific antigen (PSA), Gleason score (GS), and disease stage. Furthermore, recurrence is believed to be caused by clinically undetectable amounts of micrometastatic prostate cancer (1). To predict recurrence after treatment accurately, an improved method for detecting micrometastatic prostate cancer is required. Several means of detecting prostate cancer cells in blood have been used during investigations on the molecular staging of prostate cancer, and one of these techniques, namely, reverse transcriptase-polymerase chain reaction (RT-PCR), has been used on several occasions to detect circulating prostate cells (2, 3).
Prostate specific membrane antigen (PSMA) is a 750-amino-acid, type 2 transmembrane glycoprotein expressed on prostate epithelial cells, and has been shown to be highly prostate specific and to be expressed in a disease progression dependent manner (4-8). Many studies have been performed to determine the predictive value of PSMA as a predictor for accurate pathologic stage, but their results are still inconclusive (9-11). Furthermore, studies on the prognosis of prostate cancer following surgery according to PSMA detection are limited (8, 12).
In the present study, we investigated whether the presence of PSMA mRNA in blood has predictive value for biochemical recurrence (BCR) after radical prostatectomy.
One hundred and thirty four patients scheduled to receive radical prostatectomy for prostate cancer at our institution from March 2005 to October 2008 were prospectively enrolled. Eligible patients were required to have histologically proven adenocarcinoma of the prostate; patients that previously undergone hormonal or radiation therapy were excluded. The Institutional Review Board of the Korean National Cancer Center approved the study protocol (NCCNCS 05-049) and informed consent was obtained from all patients for the collection of clinical data, and for serum and tissue samples. Patients underwent radical prostatectomy and standard pelvic lymph node dissection. Postoperatively, no adjuvant hormonal or radiotherapy was performed until BCR had developed.
Peripheral blood samples were obtained preoperatively. Nucleated cell fractions were isolated from 5 mL whole blood samples (anti-coagulated with EDTA [ethylenediaminetetracetic acid]) by Percoll gradient centrifugation (Amersham Biosciences, Uppsala, Sweden). RNA extraction and reverse transcription were performed as described in a previous report (13).
All primers used in this study were custom designed by Bioneer (Daejeon, Korea). The intron spanning primer pairs specific for human PSMA were:- sense, 5'-CAG ATA TGT CAT TCT GGG AG GTC-3'; antisense, 5'-AAC ACC ATC CCT CCT CGA ACC-3'. For nested PCR, the sense and antisense primers were replaced by 5'-CCT AAC AAA AGA GCT GAA AAG CCC-3' and 5'-ACT GTG ATA CAG TGG ATA GCC GCT-3' respectively. The housekeeping gene β-actin was used as an internal control. PCR reactions were performed in a total volume of 20 µL containing 1 µL of RT product, 1 µM of sense and 1 µM of antisense primer, 10 mM Tris-HCl (pH 9.0), 30 mM KCl, 1.5 mM MgCl2, 0.25 mM of each dNTP, and 1 U Taq-DNA polymerase (Bioneer, Daejeon, Korea). Amplification of cDNA (0.5 µg) was performed using a tube-controlled thermal cycler (MJ Research, Waltham, MN, USA). For nested PCR, 2 µL of PCR products were amplified for an additional 40 cycles using the same reagents, but original primers were replaced with nested primers.
RT-PCR assay sensitivity was determined by assaying dilutions of LNCaP human prostate cancer cells in peripheral blood mononuclear cell (PBMC) suspensions prepared from normal donors. In our previous study, it was found that the mRNAs of 10 LNCaP cells diluted in 107 PBMCs from healthy donors could be detected by nested RT-PCR for PSMA. In the present study, all samples subjected to qualitative RT-PCR for PSMA mRNA were assayed several times to confirm test reproducibility, and results for individual samples were only subjected to further analysis after initial test results (positive or negative) had been confirmed by retesting.
PCR products (10 µL) premixed with loading dye in the PCR kit (Bioneer) and loaded into 2% agarose gel in TBE buffer (0.1 M Tris [pH 8.4], 90 mM boric acid, 1 mM EDTA), and subjected to electrophoresis for 60 min. Gels were stained with ethidium bromide and amplicons were visualized using a UV-transilluminator. To ensure internal consistency, all RT-PCR assay results were scored by two investigators unaware of clinical or pathologic data.
Complete transverse sections were taken from the apex to base of excised prostrates at 4 mm intervals. All excised prostates were examined by a single pathologist and staged according to the TNM system of the International Union Against Cancer. Gleason scores, the presence of extracapsular extension, and evidence of seminal vesicle invasion were documented.
All 134 patients were scheduled for a digital rectal examination and a serum PSA evaluation every 3 months for the first two postoperative years, biennially during the third to the fifth years, and annually thereafter. BCR was defined as a sustained PSA elevation of >0.4 ng/mL on two or more occasions. Clinical outcome analysis was performed using the NCC prostate cancer database of clinical and pathologic data.
Initially, Fisher's exact test was used to determine whether PSMA positivity was related to clinical factors. BCR-free survival curves were generated separately using the Kaplan-Meier method for patients with positive and negative RT-PCR results, and differences between the two curves were evaluated using the log-rank test. To identify factors capable of predicting BCR, univariate and multivariate recurrence analyses were performed using the Cox proportional hazards regression model. Statistical significance was accepted for P values of <0.05. All analyses were performed using the SPSS software package (version 12.0 for Windows 2000®, SPSS, Inc., Chicago, IL, USA).
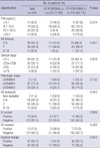
Mean patient age of the 134 patients was 60.3 years (range 49 to 70). Preoperatively, initial preoperative serum PSA levels ranged from 1.9 to 40.0 ng/mL (mean: 10.4 ng/mL, median: 7.8 ng/mL). In terms of biopsy Gleason scores, 86 patients (64.2%) had low grade (GS ≤6) disease and 48 (35.8%) high grade (GS ≥7) disease. 128 patients (94%) showed clinically prostate-confined disease (≤cT2) and the other 8 (6%) extraprostatic disease (≥cT3). With regard to pathologic stage, prostate-confined disease (≤pT2) was encountered in 98 (73.9%) patients and pT3 disease in 35 (26.1%) patients. No patients were found to have lymph node or distant metastasis. Clinical and pathological data before and after surgery are listed in Table 1.
PSMA-mRNA was detected in 24 (17.9%) patients by RT-PCR, i.e., in 14 of 97 patients (14.4%) with pT2 disease and in 9 of 35 patients (25.7%) with pT3 disease. Patients with extraprostatic extension (pT3) tended to have a PSMA mRNA positive status, but this was not statistically significant (P=0.192). With regard to the relationship between PSMA positivity and other clinical parameters, the presence of PSMA was not found to be related to serum PSA, Gleason score, or lymphovascular (LV) emboli (Table 1).
Follow-up data were available for all 134 patients with a median follow-up of 20 months (range, 3 to 46 months). BCR developed in 15 patients (11.2%) and median time to BCR was 7 months (range: 3-25 months). Of the 15 patients that developed BCR, 13 underwent hormone therapy, 1 patient is being observed using PSA check-ups, and the other 1 underwent salvage external beam radiation therapy. No deaths occurred during the study period. Kaplan-Meier analysis revealed a significant difference between those positive or negative for PSMA in terms of recurrence-free actuarial probability (log rank P=0.004, Fig. 1). Cox regression hazard analysis showed that positivity for PSMA mRNA (HR: 3.697, 95%CI 1.285-10.634, P=0.015) and a biopsy GS of ≥7 (HR: 4.500, 95%CI 1.419-14.274, P=0.011) independently predicted BCR. On the other hand, in terms of postoperative predictors of BCR, only pathologic T3 stage (HR: 4.016, 95%CI 1.194-13.511, P=0.025) was found to be significant (Table 2).
The success of radical prostatectomy is dependent on the confinement of the cancer to the prostrate. Pretreatment PSA level, biopsy GS, and clinical stage have been considered to be preoperative predictors of organ confinement. However, clinical stage based on digital rectal and radiologic examinations, is inadequate for predicting the presence of extraprostatic extension (14). Furthermore, biopsy-based Gleason scores are unsuitable predictors, because cancers graded by biopsy often differ from final surgical specimen findings (15). Moreover, pretreatment PSA levels may be dependent on benign conditions, such as, benign prostatic hyperplasia and inflammation in addition to tumor burden (16). Therefore, new strategies that can improve the accuracy of tumor staging are urgently required to identify appropriate candidates for radical prostatectomy.
Regarding studies on the molecular staging of prostate cancer, many researchers have attempted to detect circulating cancer cells in peripheral blood or bone marrow using molecular techniques (2, 3). In particular, RT-PCR is a potentially powerful molecular tool for detecting small numbers of cancer cells, and can detect tumor specific marker-expressing cells that are otherwise undetectable. In terms of molecular markers specific for prostate cancer, PSA, prostate stem cell antigen (PSCA), PSMA, and human kallikrein-2 (hK2) have been investigated (17-19).
PSMA is a typical cell-surface marker of prostate cancer, and is an integral type 2 membrane protein that is abundantly and almost universally expressed in prostate carcinoma (4-6). Based on studies, which included patients with metastases, the rate of PSMA detection in blood by RT-PCR ranges from 39 to 91%, and in patients with localized prostate cancer, PSMA positivity ranges from 10 to 72% (10, 20-22). In the present study, PSMA-mRNA was detected in 14.4% with pT2 disease and in 25.7% with pT3 disease. The wide range of reported positivities is probably due to different patient characteristics and RT-PCR assay sensitivities. With regard to relationship between RT-PCR findings and clinical parameters, several studies have failed to reveal a correlation between PSMA positivity and pathologic stage, GS, or serum PSA level, which concurs with our observations (10, 20-22).
Theoretically, PSMA detection by RT-PCR in patients with localized prostate cancer suggests the dissemination of potential micrometastatic disease, which cannot be detected by even postoperative pathologic analysis. However, previous studies have focused on determining the accuracies of RT-PCR results in terms of predicting pathologic stage in patients with prostate cancer (9-11), and to our knowledge, studies conducted over the past 10 yr on the use of molecular tools, such as, RT-PCR, for accurate pathologic staging have, as yet, failed to yield consistent results. However, given that pathologic stage and GS in surgical specimens are well-established prognostic factors of recurrence after surgery, we undertook to evaluate the predictive value of circulating tumor cells preoperatively in patients scheduled to undergo definitive treatment, such as surgery or radiation therapy. Few reports have presented relations between RT-PCR and prognostic value, such as, recurrence-free survival, and the role of RT-PCR in this context has not been determined (8, 12). Our finding that the presence of PSMA mRNA in peripheral blood can predict BCR after radical prostatectomy in patients with prostate cancer has substantial practical implications. Moreover, our results strongly indicate that PSMA detection in peripheral blood can predict disease recurrence after surgery, and show that nested RT-PCR for PSMA is a more potent predictive parameter than clinical parameters, such as, preoperative PSA level, biopsy GS, and clinical stage. Accordingly, our findings suggest that RT-PCR assays for PSMA could help identify appropriate patients for radical prostatectomy. Furthermore, a positive RT-PCR PSMA status was found to be a potent independent postsurgical predictor of recurrence by multivariate analysis incorporating postsurgical parameters, such as, pathological stage, GS, LV tumor emboli, and a positive surgical margin (RT-PCR PSMA [HR: 3.887, 95%CI 1.284-11.771, P=0.016] stage ≥pT3 [HR: 2.254, 95%CI 0.915-11.052, P=0.069]). Finally, our findings also suggest that PSMA status should be closely followed to detect localized disease recurrence.
The present study has several limitations that require consideration. First, it is controversial as to whether circulating tumor cells always cause recurrence. Therefore, additional follow-up is required to assess the significance of circulating prostate cancer cells in peripheral blood. Second, over a median follow-up of 20 months (range, 3 to 46 months), BCR developed in our cohort in less than 25 months (median 7 months, range 3 to 25), which is too short a follow-up to enable us to comment on long-term results or on delayed BCR development.
In conclusion, PSMA mRNA detection by RT-PCR in peripheral blood can be a potent preoperative predictor of BCR after radical prostatectomy.
Figures and Tables
 | Fig. 1Kaplan-Meier plot of the likelihood of RT-PCR PSMA positive cases remaining free of biochemical recurrence after radical prostatectomy (log rank P=0.0039).
PSMA, prostate specific membrane antigen; BCR, biochemical recurrence.
|
References
1. Mejean A, Vona G, Nalpas B, Damotte D, Brousse N, Chretien Y, Dufour B, Lacour B, Brechot C, Paterlini-Brechot P. Detection of circulating prostate derived cells in patients with prostate adenocarcinoma is an independent risk factor for tumor recurrence. J Urol. 2000. 163:2022–2029.

2. Zippelius A, Pantel K. RT-PCR-based detection of occult disseminated tumor cells in peripheral blood and bone marrow of patients with solid tumors. An overview. Ann N Y Acad Sci. 2000. 906:110–123.

3. Moreno JG, Croce CM, Fischer R, Monne M, Vihko P, Mulholland SG, Gomella LG. Detection of hematogenous micrometastasis in patients with prostate cancer. Cancer Res. 1992. 52:6110–6112.
4. Israeli RS, Powell CT, Fair WR, Heston WD. Molecular cloning of a complementary DNA encoding a prostate-specific membrane antigen. Cancer Res. 1993. 53:227–230.
5. Kawakami M, Nakayama J. Enhanced expression of prostate-specific membrane antigen gene in prostate cancer as revealed by in situ hybridization. Cancer Res. 1997. 57:2321–2324.
6. Troyer JK, Beckett ML, Wright GL Jr. Detection and characterization of the prostate-specific membrane antigen (PSMA) in tissue extracts and body fluids. Int J Cancer. 1995. 62:552–558.
7. Wright GL Jr, Grob BM, Haley C, Grossman K, Newhall K, Petrylak D, Troyer J, Konchuba A, Schellhammer PF, Moriarty R. Upregulation of prostate-specific membrane antigen after androgen-deprivation therapy. Urology. 1996. 48:326–334.

8. Olsson CA, De Vries GM, Benson MC, Raffo A, Buttyan R, Cama C, O'Toole K, Katz AE. The use of RT-PCR for prostate-specific antigen assay to predict potential surgical failures before radical prostatectomy: molecular staging of prostate cancer. Br J Urol. 1996. 77:411–417.

9. Grasso YZ, Gupta MK, Levin HS, Zippe CD, Klein EA. Combined nested RT-PCR assay for prostate-specific antigen and prostate-specific membrane antigen in prostate cancer patients: correlation with pathological stage. Cancer Res. 1998. 58:1456–1459.
10. Zhang Y, Zippe CD, Van Lente F, Klein EA, Gupta MK. Combined nested reverse transcription-PCR assay for prostate-specific antigen and prostate-specific membrane antigen in detecting circulating prostatic cells. Clin Cancer Res. 1997. 3:1215–1220.
11. Katz AE, Olsson CA, Raffo AJ, Cama C, Perlman H, Seaman E, O'Toole KM, McMahon D, Benson MC, Buttyan R. Molecular staging of prostate cancer with the use of an enhanced reverse transcriptase-PCR assay. Urology. 1994. 43:765–775.

12. Okegawa T, Nutahara K, Higashihara E. Preoperative nested reverse transcription-polymerase chain reaction for prostate specific membrane antigen predicts biochemical recurrence after radical prostatectomy. BJU Int. 1999. 84:112–117.

13. Joung JY, Yang SO, Jeong IG, Han KS, Seo HK, Chung J, Park WS, Lee KH. Reverse transcriptase-polymerase chain reaction and immunohistochemical studies for detection of prostate stem cell antigen expression in prostate cancer: potential value in molecular staging of prostate cancer. Int J Urol. 2007. 14:635–643.

14. Grossfeld GD, Chang JJ, Broering JM, Li YP, Lubeck DP, Flanders SC, Carroll PR. Under staging and under grading in a contemporary series of patients undergoing radical prostatectomy: results from the Cancer of the Prostate Strategic Urologic Research Endeavor database. J Urol. 2001. 165:851–856.

15. Donohue JF, Bianco FJ Jr, Kuroiwa K, Vickers AJ, Wheeler TM, Scardino PT, Reuter VA, Eastham JA. Poorly differentiated prostate cancer treated with radical prostatectomy: long-term outcome and incidence of pathological downgrading. J Urol. 2006. 176:991–995.

16. Eastham JA, Riedel E, Scardino PT, Shike M, Fleisher M, Schatzkin A, Lanza E, Latkany L, Begg CB. Variation of serum prostate-specific antigen levels: an evaluation of year-to-year fluctuations. JAMA. 2003. 289:2695–2700.
17. Su SL, Boynton AL, Holmes EH, Elgamal AA, Murphy GP. Detection of extraprostatic prostate cells utilizing reverse transcription-polymerase chain reaction. Semin Surg Oncol. 2000. 18:17–28.

18. Olsson CA, de Vries GM, Buttyan R, Katz AE. Reverse transcriptase-polymerase chain reaction assays for prostate cancer. Urol Clin North Am. 1997. 24:367–378.

19. de la Taille A, Olsson CA, Buttyan R, Benson MC, Bagiella E, Cao Y, Burchardt M, Chopin DK, Katz AE. Blood-based reverse transcriptase polymerase chain reaction assays for prostatic specific antigen: long term follow-up confirms the potential utility of this assay in identifying patients more likely to have biochemical recurrence (rising PSA) following radical prostatectomy. Int J Cancer. 1999. 84:360–364.

20. Israeli RS, Miller WH Jr, Su SL, Powell CT, Fair WR, Samadi DS, Huryk RF, DeBlasio A, Edwards ET, Wise GJ, Heston WD. Sensitive nested reverse transcription polymerase chain reaction detection of circulating prostatic tumor cells: comparison of prostate-specific membrane antigen and prostate-specific antigen-based assays. Cancer Res. 1994. 54:6306–6310.
21. Sokoloff MH, Tso CL, Kaboo R, Nelson S, Ko J, Dorey F, Figlin RA, Pang S, deKernion J, Belldegrun A. Quantitative polymerase chain reaction does not improve preoperative prostate cancer staging: a clinicopathological molecular analysis of 121 patients. J Urol. 1996. 156:1560–1566.

22. Loric S, Dumas F, Eschwege P, Blanchet P, Benoit G, Jardin A, Lacour B. Enhanced detection of hematogenous circulating prostatic cells in patients with prostate adenocarcinoma by using nested reverse transcription polymerase chain reaction assay based on prostate-specific membrane antigen. Clin Chem. 1995. 41:1698–1704.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download