Abstract
Blowflies, especially species belonging to the subfamily Luciliinae, are the first insects to lay eggs on corpses in Korea. Fast and accurate species identification has been a key task for forensic entomologists. Because conventional morphologic identification methods have many limitations with respect to forensic practice, molecular methods have been proposed to identify fly species of forensic importance. To this end, the authors amplified and sequenced the full length of the cytochrome c oxidase subunit I (COI) gene of the Luciliinae fly species collected in Korea. The results showed the COI sequences are instrumental in identifying Luciliinae fly species. However, when compared with previously reported data, considerable inconsistencies were noted. Hemipyrellia ligurriens data in this study differed significantly from two of the five pre-existing data. Two closely related species, Lucilia illustris and Lucilia caesar, showed an overlap of COI haplotypes due to four European sequences. The results suggest that more individuals from various geographic regions and additive nuclear DNA markers should be analyzed, and morphologic identification keys must be reconfirmed to overcome these inconsistencies.
Estimation of the postmortem interval (PMI) is an essential component in investigations of unnatural death (1). Before a corpse begins to decompose, the PMI can be estimated by observation of early postmortem changes, such as body cooling, muscle rigidity, and postmortem lividity. These early postmortem changes are the results of physical or chemical consequences, which do not alter the overall consistency of the corpse. However, because the early postmortem changes are disrupted by late postmortem changes, such as putrefaction and autolysis, which gradually degrade the corpse, estimation of the PMI in a decomposing body has been a difficult task in forensic practice (1). Tiny insects gathering around the corpse can provide an alternative clue because the growth rates and the occurrence of insects are dependent on the ambient temperature and the stages of decomposition, respectively (2). In most developed countries, forensic entomology, which applies insect data to the forensic investigation, is now a major approach used to estimate PMI after decomposition occurs.
Because different insect species have different growth rates and different geographic or climatic distributions, accurate species identification is critical in the practice of forensic entomology (3-6). However, species identification of insects of forensic importance, especially flies, is a very complicated and difficult process, in which the small anatomic identification keys are observed with a stereomicroscope (7). Moreover, most insect specimens recovered from the scene are immature stages, such as eggs, larvae, or pupae, which are in many instances indistinguishable among different species (8). For accurate and convenient species identification, DNA-based methods have been developed, usually analyzing the mitochondrial cytochrome c oxidase I (COI) gene or the ribosomal internal transcribed spacer 2 (ITS 2) (9-11). Because DNA sequences are divergent among different species, species identification is possible with the DNA sequence data. Currently, COI gene sequences are used as a main molecular identification tool for insects (12-14).
The subfamily Luciliinae is one of the major groups in the Family Calliphoridae (7). However, there have been limited studies about COI sequences of Korean Luciliinae fly species (15, 16). Furthermore, full-length sequences of COI genes of the Korean Luciliinae fly species have not yet been determined. The current study is the first to determine the full-length sequences of COI genes of Korean Luciliinae flies, which the authors believe, are the most important necrophagous fly species, from late spring to early fall, in Korea.
Flies were collected from various locations in Seoul, Korea. Morphologic identification was performed under a dissecting microscope by a taxonomic expert in dipterology, following procedures published by Kano and Shinonaga (7). Four individuals of Lucilia caesar, nine Lucilia illustris, two Hemipyrellia ligurriens, five Phaenicia sericata, and one Lucilia ampullacea were examined (Table 1).
After the compound eyes were removed, the whole bodies were ground to a fine powder using liquid nitrogen. Four hundred µL of lysis buffer (100 mM NaCl, 10 mM Tris-HCl, 25 mM EDTA, and 0.5% SDS [pH 8.0]) and 10 µL of proteinase K (20 µg/µL) were added to the powder and the mixture was incubated at 56℃ for 3 hr. DNA was extracted from the mixture using a conventional phenol/chloroform/isoamylalcohol method (23).
Two new primers, which span the whole COI region, were selected from the highly conserved tRNA-cystein and cytochrome c oxidase subunit II (COII) regions of the mitochondrial sequences of Chrysomya putoria (NCBI accession number NC002697), Cochliomyia hominivorax (NC002660), and Haematobia irritans (NCBI accession number NC007102). For amplification failures, several internal primers were adopted from other authors' literatures (24-26). The primer sequences are listed in Table 2.
2.5 µL of 10× Gold Buffer (Applied Biosystems, Foster City, CA, U.S.A.), 62.5 nM MgCl2, 5 nM dNTP mix (each), 5 pM primers, and 0.5 U AmpliTaq Gold DNA polymerase (Applied Biosystems) were admixed in a 25 µL total volume. After 1 cycle at 95℃ for 11 min of hot start enzyme activation, COI genes were amplified by 35 cycles at 95℃ for 30 sec, annealing temperature for 30 sec, 72℃ for 90 sec, and 72℃ for 15 min of final extension with GeneAmp 2720 or 9600 PCR Systems (Applied Biosystems). The annealing temperature was optimized for each reaction (55 to 45℃). For sequencing reaction, the residual dNTP and primers in the mixture were processed by incubation at 37℃ for 45 min with 10 U of calf intestinal phosphatase and 10 U of exonuclease I (New England BioLabs Inc., Ipswich, MA, U.S.A.). The Big-Dye Terminator, v1.1 or v3.1, Kit (Applied Biosystems) was used for DNA sequencing reaction. Sequencing reaction products were purified following the manufacturer's manual and analyzed by automatic DNA sequencers (ABI PRISM 3730xl genetic analyzer or an ABI PRISM 310 genetic analyzer).
After sequence alignment, phylogenetic analysis was performed using the neighbor-joining method with a p-distance model and 1,000 replicates of bootstrapping using MEGA 4 software (27). A COI sequence of Calliphora vicina (AJ417702) was used as the outgroup. Calculations for percent nucleotide distances were performed using MEGA4 software (27).
Twenty-one individual flies from three genera and five species in total were examined. The distance matrices of Korean Luciliinae flies are demonstrated in Table 3. The maximum intraspecific distances were 0.2, 0.0, 0.8, and 0.4% for P. sericata, H. ligurriens, L. caesar, and L. illustris, respectively. Because only one individual L. ampullacea was available, intraspecific variation in this species was not estimated. Interspecific distance was the lowest between L. caesar and L. illustris (1.4-1.9%) and the highest between L. ampullacea and H. ligurriens (8.1%). A phylogenetic tree generated from COI sequences of Korean Luciliinae flies showed no paraphyly within the genera or species levels (Fig. 1). Because there was no overlap between interspecific and intraspecific distances, the five Korean Luciliinae fly species examined in this study were distinguishable from each other by comparing COI nucleotide sequences.
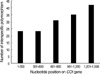
The number of polymorphic sites was 192 for total data. After intraspecific polymorphic sites were excluded, 155 polymorphic sites remained. These 155 sites are distributed a bit more in the downstream region of the COI gene (Fig. 2). This richness of interspecific polymorphism in the downstream region may suggest, in Luciliinae fly species, DNA barcoding, which spans the upstream region, may be not as useful as in other animal taxa. However, the best region choice for species identification may be controversial, because there have been no large-scale studies to investigate the whole length of COI genes pertaining to the variable Luciliinae species of forensic importance.
Table 4 demonstrates intraspecific COI nucleotide distances from Korean Luciliinae fly species and their previously reported conspecifics. The intraspecific sequence distances among H. ligurriens are outstanding (0-8.6%). A separate distance matrix for this species (data not shown) reveals these unusually high intraspecific distances are thoroughly due to two Taiwanese sequences (AY097334 and DQ453493), both morphologically inspected by Chen et al. (17, 19). These two Taiwanese sequences showed 5.5 to 8.6% distances from the conspecifics of other countries. Moreover, these two Taiwanese sequences are unusually similar to those of the Hawaiian Phaenicia cuprina (sequence distance 0.4-0.6%), which is a closely related sister species of P. sericata. Two closely related sister species, L. illustris and L. caesar showed 2.8% and 2.5% of intraspecific distances, respectively. These relatively high intraspecific distances are due to four European sequences (two L. illustris AJ551445 and EU418574, and two L. caesar AJ 417703 and DQ453488). In a phylogenetic analysis, these four sequences caused species-level paraphylies by clustering with their sister species (phylogenetic tree not shown). P. sericata and L. ampullacea showed 0-0.3% intraspecific distances.
Species-level paraphyly or even polyphyly in mitochondrial DNA, is not uncommon in the Animal Kingdom. According to Funk and Omland (28), at least 702 arthropod species are polyphyletic (26.5%). For these polyphylies, several explanations have been offered, which consist of inadequate sequence data, inaccurate taxonomy, interspecific hybridization or introgression, and incomplete sorting. In another study by Cognato (29), the intraspecific percent sequence distance of the COI genes in a lice species, Columbicola macrourae, was the highest (26.0%) among 26 insect species. However, this extraordinary intraspecific percent distance may be due to parasitic inhabitation of the louse (30). According to Cognato (29), intraspecific percent distances of the COI genes in 8 fly species (Order: Diptera) range from 0.04 to 3.5%. Therefore, intraspecific percent distances in H. ligurriens, 0 to 8.6% (in the Results section), are quite unusual among fly species. Although Wells et al. (19) insisted that validity of the genus Hemipyrellia is highly questionable, in our analysis, all sequences of H. ligurriens, except for two Taiwanese sequences, show distances within 3.6%. Further investigations, by other taxonomists, into the Taiwanese H. ligurriens formerly identified by Chen et al. (17), could ascertain the level of applicability, substantiate findings, and answer questions regarding the unusual COI haplotype.
L. illustris and L. caesar, which showed overlaps between intra- and interspecific sequence distances, are closely related sister species. Wells et al. (19) also noted a paraphyly between these two species and indicated, because L. caesar does not exist in the New World, identification of L. illustris in North America would be possible. Despite this paraphyly, because mixed clades occurred only in European strains, Asian strains are possibly distinguishable with COI sequence analyses. However, more sequence collection from various Asian and European regions would be helpful to prevent possible misidentification and to reinforce the validity of paraphyletic European sequences. Harvey et al. (20) insisted that an alternative gene choice, perhaps nuclear markers like ITS regions, may resolve COI para/polyphylies in some Calliphoridae taxa, especially when it is caused by incomplete lineage sorting. Regarding L. caesar and illustris, alternative gene analyses, including nuclear genes, may be applicable.
In conclusion, the Korean Luciliinae fly species are identifiable with COI sequence analyses. However, because of the inconsistent COI haplotypes of H. ligurriens, L. caesar, and L. illustris in a worldwide-scale comparison, more individuals from various geographic regions and alternative nuclear genes, for these three species, should be investigated.
Figures and Tables
Fig. 1
Phylogenetic analysis of Korean Luciliinae fly species. The letters Se, Am, Ca, Il, Li and Vi stand for Phaenicia sericata, Lucilia ampullacea, L. caesar, L. illustris, Hemipyrellia ligurriens, and Calliphora vicina, respectively. Calliphora vicina is included as the outgroup.

Table 3
Percent distance matrices of COI nucleotide sequences of Luciliinae flies collected in Korea

*The letters Se, Li, Ca, Il, and Am represent Phaenicia sericata, Hemipyrellia ligurriens, Lucilia caesar, Lucilia illustris, and Lucilia ampullacea, respectively. AJ417702 is a COI sequence of Calliphora vicina (outgroup). Numbers in bold indicate intraspecific distances. The intraspecific distance for Lucilia ampullacea is not available because there is only one sequence. Sequence lengths are all 1,539.
ACKNOWLEDGMENTS
The authors thank Mr. Jong Hyun Nam of Glami Co., LTD. for financial support for our study and Dr. Soochin Cho of the University of Michigan for a phylogenetic advice on this manuscript.
References
1. Saukko PJ, Knight B. Knight's forensic pathology. 2004. 3rd ed. London, New York: Arnold;52–97.
2. Gennard DE. Forensic entomology: an introduction. 2007. Chichester, England; Hoboken, NJ: John Wiley & Sons;115–130.
3. Anderson GS. Minimum and maximum development rates of some forensically important Calliphoridae (Diptera). J Forensic Sci. 2000. 45:824–832.

4. Kamal AS. Comparative study of thirteen species of sarcosaprophagous Calliphoridae and Sarcophagidae (Diptera) I. Bionomics. Ann Entomol Soc Am. 1958. 51:261–271.
5. Marchenko MI. Medicolegal relevance of cadaver entomofauna for the determination of the time of death. Forensic Sci Int. 2001. 120:89–109.

6. Nishida K. Experimental studies on the estimation of postmortem intervals by means of fly larvae infesting human cadavers. Nihon Hoigaku Zasshi. 1984. 38:24–41.
7. Kano R, Shinonaga S. Calliphoridae (Insecta: Diptera). 1968. 1st ed. Tokyo: Biological [i.e. Biogeographical] Society of Japan.
8. Wells JD, Sperling FA. Molecular phylogeny of Chrysomya albiceps and C. rufifacies (Diptera : Calliphoridae). J Med Entomol. 1999. 36:222–226.
9. Sperling FA, Anderson GS, Hickey DA. A DNA-based approach to the identification of insect species used for postmortem interval estimation. J Forensic Sci. 1994. 39:418–427.
10. He L, Wang S, Miao X, Wu H, Huang Y. Identification of necrophagous fly species using ISSR and SCAR markers. Forensic Sci Int. 2007. 168:148–153.

11. Wells JD, Williams DW. Validation of a DNA-based method for identifying Chrysomyinae (Diptera: Calliphoridae) used in a death investigation. Int J Legal Med. 2007. 121:1–8.

12. Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003. 270:Suppl 1. S96–S99.

13. Rubinoff D. Utility of mitochondrial DNA barcodes in species conservation. Conserv Biol. 2006. 20:1026–1033.
14. Shneyer VS. On the species-specificity of DNA: fifty years later. Biochemistry (Mosc). 2007. 72:1377–1384.

15. Zhang Y, Park SH, Yu DH, Yu GY, Jung HJ, Jo TH, Hwang JJ. Comparison of Mitochondrial Cytochrome Oxidase Subunit I (CO I) Sequences of Five Blow Fly Species in Korea. Korean J Leg Med. 2006. 30:154–159.
16. Zhang Y, Park SH, Yu DH, Yu GY, Jung HJ, Jo TH, Hwang JJ. Identification Blow Fly Species in Korea by Mitochondrial DNA 'Barcodes'. Korean J Leg Med. 2007. 31:51–58.
17. Chen WY, Hung TH, Shiao SF. Molecular identification of forensically important blow fly species (Diptera: Calliphoridae) in Taiwan. J Med Entomol. 2004. 41:47–57.

18. Wallman JF, Leys R, Hogendoorn K. Molecular systematics of Australian carrion-breeding blowflies (Diptera: Calliphoridae) based on mitochondrial DNA. Invertebr Syst. 2005. 19:1–15.
19. Wells JD, Wall R, Stevens JR. Phylogenetic analysis of forensically important Lucilia flies based on cytochrome oxidase I sequence: a cautionary tale for forensic species determination. Int J Legal Med. 2007. 121:229–233.

20. Harvey ML, Gaudieri S, Villet MH, Dadour IR. A global study of forensically significant calliphorids: implications for identification. Forensic Sci Int. 2008. 177:66–76.

21. Stevens JR, Wall R, Wells JD. Paraphyly in Hawaiian hybrid blowfly populations and the evolutionary history of anthropophilic species. Insect Mol Biol. 2002. 11:141–148.

22. Stevens JR. The evolution of myiasis in blowflies (Calliphoridae). Int J Parasitol. 2003. 33:1105–1113.

23. Butler JM. Forensic DNA typing: biology & technology behind STR markers. 2001. San Diego, CA; London: Academic Press;33–62.
24. Harvey ML, Dadour IR, Gaudieri S. Mitochondrial DNA cytochrome oxidase I gene: potential for distinction between immature stages of some forensically important fly species (Diptera) in western Australia. Forensic Sci Int. 2003. 131:134–139.

25. Harvey ML, Mansell MW, Villet MH, Dadour IR. Molecular identification of some forensically important blowflies of southern Africa and Australia. Med Vet Entomol. 2003. 17:363–369.

26. Saigusa K, Takamiya M, Aoki Y. Species identification of the forensically important flies in Iwate prefecture, Japan based on mitochondrial cytochrome oxidase gene subunit I (COI) sequences. Leg Med (Tokyo). 2005. 7:175–178.

27. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. 24:1596–1599.

28. Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Systemat. 2003. 34:397–423.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download