Abstract
Since vascular endothelial growth factor (VEGF) is known to be a potent pro-angiogenic factor, we evaluated the potential association of two VEGF gene polymorphisms (-634G>C and 936C>T) with the susceptibility and the clinicopathologic characteristics of colorectal cancer (CRC). The VEGF genotypes were determined using fresh colorectal tissue from 465 patients who had undergone a surgical resection and peripheral blood lymphocytes from 413 healthy controls by PCR/DHPLC assay. For the -634G>C polymorphism, the -634 GC or CC genotype was associated with a decreased risk of CRC (odds ratio [OR], 0.62; p=0.001) as a dominant model of C allele, whereas the 936 TT genotype correlated with advanced stage/ metastasis, a high serum level of CA19-9, and an higher grade in patients with CRC. In the haplotype analyses, haplotype -634C/936C and -634G/936T were associated with a decreased susceptibility of CRC (OR, 0.53 and 0.56; p<0.001, respectively). These observations imply that the VEGF gene polymorphisms may be associated with the susceptibility or clinicopathologic features of CRC. However, further studies of other VEGF sequence variants and their biological functions are needed to understand the role of the VEGF gene polymorphisms in the development and progression of CRC.
Colorectal cancer (CRC) is the second leading cause of cancer- related deaths in the U.S.A. and Europe (1). During the last few years, many attempts have been made to define the biological profile of CRC in order to improve early diagnosis and the prognostic stratification, and eventually find a cure (2, 3). Although many biological factors have been implicated in the development of CRC, a clinical relevance has not yet been reached for most of them.
Angiogenesis, the formation of a new blood vessel from endothelial precursors, is a prerequisite for the development, growth, and progression of solid malignancies (4). The vascular endothelial growth factor (VEGF) is one of the most potent endothelial cell mitogens and plays a critical role in angiogenesis. VEGF specifically binds to VEGF receptor tyrosine kinase on endothelial cells to initiate intracellular signal transduction pathways that mediate angiogenesis and vascular permeability. In addition to stimulating angiogenesis, VEGF may also have autocrine functions, acting as a survival factor for tumor cells by protecting them from various forms of stress and apoptosis (5-8).
The VEGF gene is located on chromosome 6p21.3 and consists of eight exons that exhibit alternative splicing to form a family of proteins (9, 10). Several polymorphisms have been associated with variations in VEGF protein production and reported to be involved in the development of several tumors (11, 12) and autoimmune diseases (13, 14). Given these results, it is also possible that a functional genetic variation in the VEGF gene may contribute to the development and progression of CRC. However, there have been no studies in the literature that have investigated the single nucleotide polymorphisms (SNPs) of the VEGF gene and their relationship to the susceptibility of CRC. Accordingly, the present study examined 2 VEGF gene polymorphisms (-634G>C and 936C>T)previously reported to be related with VEGF production, and evaluated the potential association of three VEGF gene polymorphisms with the susceptibility and clinicopathologic characteristics of CRC in the Korean population.
All the tissues investigated in this study were obtained from 465 native Korean patients with CRC who had undergone a surgical resection between January 2003 and August 2006 at Kyungpook National University Hospital (Daegu, Korea). Based on a health-screening questionnaire, 413 unrelated, healthy Korean individuals without known medical problems were also enrolled as the control group. The study was approved by the Institutional Research Board of Kyungpook National University Hospital, and all individuals gave written informed consent for study participation.
The extraction of genomic DNA from the peripheral blood lymphocytes of the control group was performed using proteinase- K digestion and phenol/chloroform extraction. For the patient group, the genomic DNA was extracted from fresh colorectal tumor tissue at the time of surgery using a Wizard genomic DNA purification kit (Promega, Madison, WI, U.S.A.). The VEGF -634G>C and 936C>T genotypes were determined using a polymerase chain reaction/denaturing high-performance liquid chromatography (PCR/DHPLC) assay as described in the previous studies (17, 18). PCR primers were designed based on a Genbank reference sequence (accession no. NT_007592). The PCR primers used for the -634G>C and 936C>T polymorphisms were 5'-CGACGGCTTGGGGAGATTGC- 3'(forward) and 5'-GGGCGGTGTCTGTCTGTCTG- 3'(reverse); and 5'-AGGGTTTCGGGAACCAGATC- 3'(forward) and 5'-CTCGGTGATTTAGCAGCAAG- 3'(reverse), respectively. The PCR reactions were performed in a 50-µL reaction volume containing 50 ng genomic DNA, 50 pM/L each primer, 10 mM/L dNTP, 5×Q- solution, 10×PCR buffer (Tris-HCl , KCl, 15 mM/L MgCl2, [NH2] 2SO4; pH8.7) and 2.5 units of HotStarTaq polymerase (QIAGEN, Hilden, Germany). The PCR cycle conditions consisted of an initial denaturation step at 94℃ for 15 min, followed by 40 cycles of 45 sec at 94℃, 45 sec at 57℃, 45 sec at 72℃, and a final elongation at 72℃ for 10 min. The PCR products were denatured at 94℃ for 10 min, and hybridized for 45 min, and screened for a heterozygous polymorphism based on a DHPLC analysis using a gradient solution of 0.1 M TEAA (pH 7.0), 0.1 M TEAA, 25% acetonitrile, a washing solution with 8% acetonitrile (syringe washing solution), and 75% acetonitrile (DNASep® Cartridge UltraClean and Storage Solution, Transgenomic, Omaha, NE, U.S.A.), Column: alkylated nonporous poly (styrene-divinylbenzene) DNASep® Cartridge (Transgenomic, Omaha, NE, U.S.A.), flow rate: 0.9 mL/min, oven temperature: 64℃, and UV: 260 nm.
The remaining samples showing a single peak on the DHPLC were mixed with the PCR products of a known homozygous genotype (homozygous A), and hybridized to run the DHPLC again, as described above. Another type of homozygous genotype (homozygous B) was confirmed when a double peak appeared on the DHPLC. Several samples with three different patterns on the DHPLC were directly sequenced to reconfirm the accuracy of the DHPLC (Fig. 1).
The Hardy-Weinberg equilibrium for each polymorphism was analyzed using the χ2 test, which was also used to examine the statistical significance of the differences in the allele frequency and genotype distribution between the groups. The odds ratios (ORs) and 95% confidence intervals (CIs) were obtained using an unconditional logistic regression analysis. For the patient group, the χ2-test or ANOVA was used to evaluate the relation between each polymorphism and the tumor characteristics. The software PHASE (version 2.1), which uses a Bayesian statistical method, was used to reconstruct the haplotypes for the VEGF gene (19). The p value was generated using a 2-sided test. The analyses were conducted using SPSS version 12.0 (SPSS, Chicago, IL, U.S.A.) and SAS Genetic software (SAS Institute, Cary, NC, U.S.A.).
The clinical and pathologic characteristics of patients are summarized in Table 1. The median age of the patients was 64 (range, 21-89) yr, and 248 patients (51.8%) were male. There was no difference in age between the patient and control groups, while there was a male predominance in the control group. The pathologic stages after the surgical resection were as follows: stage I (n=80, 17.2%), stage II (n=157, 33.8 %), stage III (n=157, 33.8%), and stage IV (n=71, 15.3%).
The VEGF genotypes were successfully identified in all 878 enrolled subjects, and the frequencies of the genotypes and alleles are listed in Table 2. The genotype distributions of the two polymorphisms among the patients and controls followed the Hardy-Weinberg equilibrium, and the frequencies of the -634C and 936T alleles among the healthy Koreans were 0.473 and 0.209, respectively. The incidence of each genotype with the -634G>C polymorphism differed between the two groups (p=0.001), where the combined GC and CC genotype was significantly associated with a decreased risk of CRC (OR, 0.62; 95% CI, 0.47-0.83; p=0.001) compared with the GG genotype as the dominant model for the C allele. For the 936C>T polymorphism, there was no difference in the genotype or allele distribution between the patient and control groups (p=0.64 and 0.71, respectively).
For the patient group, the TT genotype of 936C>T polymorphism was significantly associated with advanced stage (OR, 3.83; 95% CI, 1.22-12.03; p=0.02), distant metastasis (OR, 8.38; 95% CI, 3.00-23.51; p<0.001), high serum level of CA19-9 (OR, 2.96; 95% CI, 1.04-8.40; p=0.04), and higher grade (p=0.002). However, no association of -634G>C polymorphism with clinicopathologic features was observed (Table 3, 4).
The VEGF -634G>C and 936C>T polymorphisms exhibited an intermediate linkage disequilibrium (D'=0.50), and Table 5 shows the frequencies and ORs of the 4 reconstructed haplotypes for the two polymorphisms of the VEGF gene, as predicted by a Bayesian algorithm. Haplotype GC was the most frequent type in the patient group (41.5%), whereas haplotype CC was most frequent in the control group (43.1%). When compared with wild type (GC), the haplotype CC (OR, 0.53; 95% CI, 0.43-0.66; p<0.001) and GT (OR, 0.56; 95% CI, 0.42-0.74; p<0.001) were significantly associated with a decreased susceptibility of CRC. However, no significant associations were observed between any of the haplotypes and the clinical features of CRC (data not shown).
The present study investigated the potential impact of 2 VEGF gene polymorphisms on the susceptibility and clinicopathologic features of colorectal adenocarcinoma in quite a large population of Korean patients. As a result, the frequencies of -634C and 936T alleles were 0.473 and 0.209, respectively, which differ from those of Japanese (0.353 and 0.150, respectively) as well as those of Caucasians (0.335 and 0.150, respectively) (20, 21). Moreover, it was observed that the combined GC and CC genotypes of the -634G>C polymorphism, and haplotype -634C/936C and -634G/936T were associated with a decreased susceptibility, while 936 TT genotype with an adverse clinicopathologic feature of CRC. Given the homogenous ethnic background of the Korean patients, any potential confounding effect due to ethnicity is likely to be small in the present study. However, these results might be caused by somatic alteration in the process of colorectal carcinogenesis because tumor tissue was used for DNA source of the patient group in the present study, although data on the difference between germline and somatic SNPs of VEGF gene have not been reported. Furthermore, the effect of the VEGF gene polymorphisms on the susceptibility of CRC may be due to linkage disequilibrium with other functional variants in the VEGF gene or other cytokine gene (22, 23).
Since VEGF or its family plays a critical role in tumor-related angiogenesis, several functional polymorphisms in the VEGF gene have already been reported to be associated with a VEGF gene expression or an increased risk of solid tumors (11, 12, 23, 24), making them potential predictive markers for clinical outcomes (22-24). For example, the -634C and 936T alleles as a dominant model or CGT haplotype (-1498T >C, -634G>C, and 936C>T) among the VEGF gene polymorphisms have been associated with a significantly decreased risk of small cell lung cancer (SCLC), while the TCC haplotype conferred a significantly increased risk of SCLC in a previous study by the present authors (25). Plus, a strong association between the -634 CC or 936 TT genotypes and a larger tumor size was observed, while the -634 CC genotypes was strongly correlated with poor differentiation and advanced stage of disease in gastric cancer (24).
One possible explanation for these results is that the DNA sequence variations in the VEGF gene may alter VEGF production and/or activity, thereby causing inter-individual differences in the development and progression of tumors. A few studies have already reported that VEGF gene polymorphisms are associated with VEGF production, yet the results are inconsistent. Awata et al. (26) reported an association between the -634 CC genotype and a higher serum VEGF concentration in the normal Japanese population. Koukourakis et al. (27) also reported that genetic polymorphisms including the -634 region were correlated with VEGF protein expression in cancer cells and angiogenesis in tumor tissue. Renner et al. (28) demonstrated that VEGF plasma levels were significantly lower in carriers of the 936T allele, whereas Krippl et al. (23) reported that the 936T allele was associated with a lower VEGF plasma level and decreased risk of breast cancer (OR, 0.51; 95% CI, 0.38-0.70). Meanwhile, Watson et al. (10) documented that the GG genotype for the -634G>C polymorphism was significantly correlated with higher VEGF production from stimulated peripheral blood mononuclear cells. Stevens et al. (29) also reported that haplotype -460C/+405G had a higher promoter activity than haplotype -460T/+405C.
In summary, the -634G>C and 936C>T polymorphisms in the VEGF gene was found to be associated with susceptibility or adverse clinicopathologic features of CRC in the current study. However, further studies of other VEGF sequence variants and their biological functions are needed to understand the role of the VEGF polymorphisms and haplotypes in the development and progression of CRC. Moreover, since genetic polymorphisms often vary between ethnic groups, more studies are also warranted to clarify the association between the VEGF polymorphisms and CRC in diverse ethnic populations.
Figures and Tables
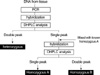 | Fig. 1Algorithm for genotyping of VEGF gene polymorphisms using a polymerase chain reaction/denaturing high-performance liquid chromatography (PCR/DHPLC) assay. |
Table 2
Genotype distributions (%) and allele frequencies in patients with colorectal cancer and controls

Table 3
Association of VEGF gene polymorphisms with clinical characteristics in patients with colorectal cancer (n=465)

References
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005. 55:74–108.

2. Pokorny RM, Hunt L, Galandiuk S. What's new with tumor markers for colorectal cancer? Dig Surg. 2000. 17:209–215.

3. Crawford NP, Colliver DW, Galandiuk S. Tumor markers and colorectal cancer: utility in management. J Surg Oncol. 2003. 84:239–248.

4. Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000. 407:242–248.

5. Tran J, Master Z, Yu JL, Rak J, Dumont DJ, Kerbel RS. A role for survivin in chemoresistance of endothelial cells mediated by VEGF. Proc Natl Acad Sci USA. 2002. 99:4349–4354.

6. Bachelder RE, Crago A, Chung J, Wendt MA, Shaw LM, Robinson G, Mercurio AM. Vascular endothelial growth factor is an autocrine survival factor for neuropilin-expressing breast carcinoma cells. Cancer Res. 2001. 61:5736–5740.
7. Harmey JH, Bouchier-Hayes D. Vascular endothelial growth factor (VEGF), a survival factor for tumour cells: implications for antiangiogenic therapy. Bioessays. 2002. 24:280–283.

8. Katoh O, Takahashi T, Oguri T, Kuramoto K, Mihara K, Kobayashi M, Hirata S, Watanabe H. Vascular endothelial growth factor inhibits apoptotic death in hematopoietic cells after exposure to chemotherapeutic drugs by inducing MCL1 acting as an antiapoptotic factor. Cancer Res. 1998. 58:5565–5569.
9. Vincenti V, Cassano C, Rocchi M, Persico G. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996. 93:1493–1495.

10. Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000. 12:1232–1235.

11. Lin CC, Wu HC, Tsai FJ, Chen HY, Chen WC. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for prostate cancer. Urology. 2003. 62:374–377.

12. Ku KT, Wan L, Peng HC, Tsai MH, Tsai CH, Tsai FJ. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for oral cancer. Oral Oncol. 2005. 41:497–502.

13. Han SW, Kim GW, Seo JS, Kim SJ, Sa KH, Park JY, Lee J, Kim SY, Goronzy JJ, Weyand CM, Kang YM. VEGF gene polymorphisms and susceptibility to rheumatoid arthritis. Rheumatology (Oxford). 2004. 43:1173–1177.

14. Salvarani C, Boiardi L, Casali B, Olivieri I, Cantini F, Salvi F, Malatesta R, La Corte R, Triolo G, Ferrante A, Filippini D, Paolazzi G, Sarzi-Puttini P, Nicoli D, Farnetti E, Chen Q, Pulsatelli L. Vascular endothelial growth factor gene polymorphisms in Behcet's disease. J Rheumatol. 2004. 31:1785–1789.
15. Hamilton SR, Aaltnen LA, editors. WHO Classification of Tumours: Pathology and Genetics of Tumours of Digestive System. 2000. Geneva, Switzerland: WHO.
16. Greene FL. TNM staging for malignancies of the digestive tract: 2003 changes and beyond. Semin Surg Oncol. 2003. 21:23–29.

17. Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: a review. Hum Mutat. 2001. 17:439–474.

18. Kosaki K, Udaka T, Okuyama T. DHPLC in clinical molecular diagnostic services. Mol Genet Metab. 2005. 86:117–123.

19. Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003. 73:1162–1169.

20. Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, Inukai K, Inoue I, Katayama S. A common polymorphism in the 5' untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002. 51:1635–1639.
21. Langsenlehner U, Wolf G, Langsenlehner T, Gerger A, Hofmann G, Clar H, Wascher TC, Paulweber B, Samonigg H, Krippl P, Renner W. Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian "tumor of breast tissue: incidence, genetics, and environmental risk factors" study. Breast Cancer Res Treat. 2008. 109:297–304.

22. McCarron SL, Edwards S, Evans PR, Gibbs R, Dearnaley DP, Dowe A, Southgate C, Easton DF, Eeles RA, Howell WM. Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res. 2002. 62:3369–3372.
23. Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, Paulweber B, Haas J, Samonigg H. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer. 2003. 106:468–471.

24. Tzanakis N, Gazouli M, Rallis G, Giannopoulos G, Papaconstantinou I, Theodoropoulos G, Pikoulis E, Tsigris C, Karakitsos P, Peros G, Nikiteas N. Vascular endothelial growth factor polymorphism in gastric cancer development, prognosis, and survival. J Surg Oncol. 2006. 94:624–630.
25. Lee SJ, Lee SY, Jeon HS, Park SH, Jang JS, Lee GY, Son JW, Kim CH, Lee WK, Kam S, Park RW, Park TI, Kang YM, Kim IS, Jung TH, Park JY. Vascular endothelial growth factor gene polymorphisms and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2005. 14:571–575.
26. Awata T, Kurihara S, Takata N, Neda T, Iizuka H, Ohkubo T, Osaki M, Watanabe M, Nakashima Y, Inukai K, Inoue I, Kawasaki I, Mori K, Yoneya S, Katayama S. Functional VEGF C-634G polymorphism is associated with development of diabetic macular edema and correlated with macular retinal thickness in type 2 diabetes. Biochem Biophys Res Commun. 2005. 333:679–685.

27. Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004. 46:293–298.

28. Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res. 2000. 37:443–448.

29. Stevens A, Soden J, Brenchley PE, Ralph S, Ray DW. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res. 2003. 63:812–816.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download