Abstract
We compared a real time-nucleic acid sequence-based amplification (RTi-NASBA) with conventional NASBA, galactomannan enzyme immunosorbent assay (GM-EIA), and Mycology Study Group of the European Organization for Research and Treatment of Cancer (EORTC/MSG) criteria for the diagnosis of invasive aspergillosis (IA). From May 2004 to May 2005, blood samples (314 in total) were collected twice a week from 78 patients with hematologic diseases during neutropenic fever after chemotherapy or hematopoietic stem cell transplantation. Results were compared with each other on the basis of EORTC/ MSG criteria. The cutoff of conventional NASBA was set to be 3.5; GM 0.5; RTi-NASBA, 20% above the negative control. There were 22 patients with IA (7 probables and 15 possibles) and 56 patients with nonfungal infection. The Kappa statistic for RTi-NASBA versus conventional NASBA was 0.80 (0.66-0.82; p<0.001) indicating that there was fairly good accordance between two tests. RTi-NASBA showed sensitivity 0.96, specificity 0.43, positive- and negative-predictive value 0.40 and 0.96, respectively. GM showed good specificity (0.98), while the sensitivity (0.45) was poor. When we use the combination of GM with either of two NASBAs, the sensitivity was improved up to 100%. In conclusion, RTi-NASBA could be a good alternative to the conventional one for the screening of IA.
The diagnosis of Aspergillus species has been a major challenge in the management of patients with invasive aspergillosis (IA). Although early detection is critical in clinical decision making, conventional blood culture rarely detects Aspergillus species (1-3). Therefore, we should rely on nonculture-based methods for the diagnosis of IA.
Several molecular detection methods have been devised (2). Of these, the detection of galactomannan by enzyme immunosorbent assay (GM-EIA) is one of the standard measures (4-6) and was included as an important component of microbiological factors in the diagnostic criteria devised by the Mycology Study Group of the European Organization for Research and Treatment of Cancer (EORTC/MSG) (7). In addition to this, we have used nucleic acid sequence-based amplification (NASBA) as a nucleic acid detection method. NASBA is an isothermal amplification process to detect mRNA by hybridization using electrochemiluminescent (ECL) probes (8). With this method, we have reported the cutoff NASBA index value for clinical diagnosis and suggested its usefulness for monitoring and management of clinical course of invasive aspergillosis for the first time (9).
Recently, this technique adopts real-time scheme (RTi-NASBA) by combining with molecular beacon probes (10). It has been applied to the diagnosis of various pathogens such as HIV (11, 12), hepatitis B virus (13), influenza virus (14), etc., but there has been only one preliminary report on the detection of Aspergillus species so far (15).
In this study, we have compared RTi-NASBA with conventional NASBA and GM-EIA for the diagnosis of IA. To our knowledge, this is the first comparative study of RTi-NASBA for the detection of Aspergillus species.
We performed this study from May 2004 to May 2005 in the Catholic hematopoietic stem cell transplantation (HSCT) center affiliated to The Catholic University of Korea, College of Medicine. Febrile neutropenic patients unresponsive to broad-spectrum antibiotics by 72 to 96 hr of initiation were enrolled consecutively into the study. Blood samples were usually obtained twice a week until recovery from neutropenia. IA was diagnosed on the basis of the standard definitions of invasive fungal infections using the EORTC/MSG criteria (10). Using these definitions, we classified the study population into four probability levels: proven, probable and possible IA, and non-fungal infection (NFI).
NASBA was performed in accordance with the scheme we used in the previous study (9). In brief, RNA was extracted from an EDTA-treated whole blood sample using NucliSens Isolation Reagents (BioMerieux Korea & Diagenex, Seoul, Korea) and QIAGEN RNeasy Minikit (Quiagen, Germany). Then we performed NASBA with a NucliSens kit and an electrochemiluminescence (ECL) detector (Bio-Merieux Korea & Diagenex, Seoul, Korea) according to the manufacturer's instructions. Primers and capture probes for detecting Aspergillus species were made using the scheme proposed by Loeffler et al. (16). The sequence of primer 1 (including the T7-promoter region) was:
5'-AATTCTAATACGACTCACTATAGGGGAGCAAAGGCCTGCTTTGAACA-3', and that of primer 2 (including ECL-tail) was:
5'-GATGCAAGGTCGCATATGAGGCCGCGGTAATTCCAGCTCCAATA-3'.
The capture probe sequence, which binds to clinically relevant Aspergillus species, was 5'-GGTCCGCCTCACCGCGAGTACTG-3'. Results are expressed as ECL counts. The wild type (WT) signal value in each experiment was set at 0.01 × the signal from the ruthenium-labeled paramagnetic beads as reference solution. Every NASBA index was calculated as the ratio: (measured ECL count of NASBA)/(WT signal value).
The initial value of the NASBA index of every patient was included in the calculation. The cutoff value of the NASBA index was determined by using receiver-operating characteristic (ROC) analysis. Sensitivity and specificity were calculated using cases of IA on the basis of EORTC/MSG criteria as a gold standard.
The primer 1 & 2 were same as those for conventional NASBA. The molecular beacon was labeled with 6-carboxyfluorescein (6-FAM) at its 5'-end and quencher DABCYL at its 3'-end. The sequence was 5'-(FAM) CGA TCG GGT CCG CCT CAC CGC GAG TAC TGC GAT CG-DABCYL-3'. RTi-NASBA was performed using NucliSens EasyQ incubator and detection system (BioMerieux Korea & Diagenex, Seoul, Korea) according to the manufacturer's instruction.
Threshold level of RTi-NASBA was determined according to the manufacturer's instruction. To determine the baseline fluorescence (BF), a random sampling of 20 specimens from healthy volunteers were analysed in four different runs using the NucliSens EasyQ Analyzer. The threshold level with real-time detection was calculated by multiplying the average baseline fluorescence (BFmean) resulting from a random sampling with the ratio (BFratio), as described in the following equation: Threshold level=BFmean × BFratio where BFratio equals to BFhighest divided by BFlowest. In this experiment, we obtained the average threshold value as 1.20 by multiplying the BF value of 1.11 with the BF ratio of 1.07, which was approximately 20% above the value of negative control.
The scheme of RTi-NASBA is illustrated in Fig. 1.
Serum GM was measured by sandwich ELISA using rat monoclonal antibody EB-A2 (Platelia Aspergillus, Bio-Rad Korea). Results are expressed as the ratio of the optical density of the sample to that of a standard sample containing 1 ng of GM/mL. A GM index of ≥0.5 in at least two consecutive samples was regarded as positive.
The result of RTi-NASBA was compared with those of NASBA-ECL and GM-EIA on the basis of EORTC/MSG criteria. We also assessed the combination of GM-EIA and NASBA-ECL or RTi-NASBA for the diagnostic yield of IA. Concordance of methods was measured using Kappa statistics.
A total of 314 samples were obtained from 22 patients with IA (7 probables and 15 possibles) and 56 patients with NFI.
The male-to-female ratio was 10:12. Most cases were presented with pneumonia (n=13), but there were one case of sinusitis, and eight cases were presented with intractable fever. Thirteen (59.1%) of the cases were diagnosed with acute myelocytic leukemia, and 5 with acute lymphocytic leukemia. There were also one case of chronic myelocytic leukemia and one myelodysplastic syndrome. All but one case received cytotoxic chemotherapy. The mortality was 23% (5/22).
Using probable and possible cases as a gold standard, the area under the ROC curve (AUC) was 0.937±0.02 (95% confidence interval. 0.902-0.972; p<0.001). The optimal inflection point was located between NASBA index 3.105 (sensitivity 0.91, specificity 0.68) and 3.559 (sensitivity 0.91, specificity 0.77), as illustrated in Fig. 2. On the basis of this analysis, we decided the cutoff value of NASBA-ECL to be 3.5.
The sensitivity of NASBA-ECL (3.5 as a cutoff point) was high (0.95) at the expense of low specificity (0.45). Positive and negative predictive values were 0.40 and 0.96, respectively (Table 1).
The sensitivity and specificity of RTi-NASBA showed a profile similar to NASBA-ECL: high sensitivity and negative predictive value (0.96 each) in contrast to low specificity and positive predictive value (0.43 and 0.40, respectively).
The sensitivity of GM-EIA (0.5 as a cutoff point) was rather low (0.45), while the specificity was high (0.98). Positive and negative predictive values were 0.91 and 0.82, respectively.
As depicted in Table 2, the agreement between two NASBA methods was fairly good with kappa statistics of 0.80 (95% confidence interval: 0.66-0.82, p<0.01).
When either of GM-EIA index ≥0.5 or NASBA-ECL ≥3.5 were used as positive cutoff values, the diagnostic yield of IA was enhanced with sensitivity up to 100%. The diagnostic power was also enhanced when either of GM-EIA index or RTi-NASBA was used as positive markers: the sensitivity also rose up to 100% (Table 3).
In addition to GM-EIA, various diagnostic measures based on the molecular detection have been developed so far (1-3). Among them, we used the amplification and detection of mRNA from Aspergillus (NASBA) and applied the method to patients with febrile neutropenia for the enhancement of diagnostic yield and establishment of treatment guideline as reported in our previous report (9), which was meaningful on the aspect of the first presentation of practical guideline for the clinical decision making.
In this study, we evaluated the RTi-NASBA as an alternative to the conventional NASBA. RTi-NASBA is reported to be advantageous over conventional one in many ways; the amplification and detection are performed in one tube and thus RTi-NASBA has fewer steps prior to amplification, no post-amplification hybridization, less time-consuming, and lower chance of amplicon contamination (17). It is also easier to perform and quantify. Although these differences are not significant between these two NASBA methods, we thought these advantages could outweigh the slightly higher reagent cost. In our study, the interface of RTi-NASBA was user-friendly, easy to perform, and less laborious than conventional NASBA. Time-saving was about 30 min; 3 hr in RTi-NASBA vs. 3.5 hr in NASBA on average.
The good measure of agreement (kappa value=0.8) suggested that RTi-NASBA could be a comparable to the conventional one with respect to the diagnostic power.
RTi-NASBA as well as conventional NASBA (NASBA-ECL) showed an excellent sensitivity and negative predictive value with poor specificity and positive predictive value, while GM-EIA showed opposite performance (good specificity with poor sensitivity).
Though it is enrolled as a formal component of diagnostic criteria in EORTC/MSG, the low sensitivity of GM-EIA limits its value as a screening measure (18). Therefore, the diagnostic yield of GM-EIA should be complemented by another method, and conventional- or RTi-NASBA could be the one as suggested in our previous report. However, the capability of these two NASBA methods could be questioned because of low specificity in this study. This can be interpreted in many ways. Which method you will prefer depends on which goal you want from the detection method: an accurate diagnosis or screening as many as possible. As the main objective of nonculture-based detection of IA is a screening for the clinical decision making rather than accurate diagnosis, higher sensitivity at the expense of specificity would be more important.
The choice of optimal cutoff value depends on the purpose of the test. As the main objective of this study was to evaluate the test for initial screening of patients with a risk of IA, we chose a value of the NASBA index giving an appropriately high sensitivity and an acceptably low false positive rate (1-specificity).
The diagnostic yield could be enhanced by combination of GM-EIA with either of conventional- or RTi-NASBA. In our study, combination of GM with either of two NASBA methods yielded the sensitivity of 100%.
Our study has some limitations. We did not include proven cases in this study. Adding the probable and possible cases without proven cases to the gold standard might have influenced the validation of our tests. Practically, obtaining a specimen for confirmation is much more difficult in almost all clinical settings because of the inability to perform tissue biopsies and autopsies owing to immunocompromised status as well as refusal of patients and families. However, we think that the high stringency of EORTC/MSG criteria for probable and possible cases could specifically select invasive fungal diseases with a very high index of suspicion. In spite of all these drawbacks, this study could offer us the validity of two NASBAs as adequate screening tools.
In summary, RTi-NASBA could be a good alternative to the conventional NASBA using the ECL detection system. If either the GM-EIA or NASBA (conventional- or RTi-NASBA) suggests IA, it could provide a valid clue to the initiation of antifungal treatment before obtaining confirmatory test results.
Adding these diagnostic markers for fungal disease to the treatment strategies could be also beneficial. For this purpose, further studies are underway to set up the quantitative measurement of RTi-NASBA using internal calibrator RNA as a control and to establish the basis of pre-emptive antifungal treatment guided by these molecular diagnostic markers.
Figures and Tables
Fig. 1
Scheme of real time-nucleic acid sequence-based amplification (RTi-NASBA); upper: Sequence of RTi-NASBA; lower left: structure of molecular beacon and the process of detection by emission of fluorescence from R (recipient) on receiving light and freed from the inhibitory action of Q (quencher); lower right: a sample of the result from RTi-NASBA.
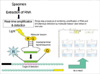
Fig. 2
Receiver-operating characteristic (ROC) curve and a table of statistics for selected thresholds for the cutoff NASBA value on the group of patients with probable and possible invasive aspergillosis as a standard (AUC: area under the ROC curve, 95% C.I.: 95% confidence interval).

Table 1
Sensitivity (sn), specificity (sp), positive- and negative-predictive value (ppv and npv) of NASBA, real time (RTi)-NASBA, and galactomannan enzyme immunosorbent assay (GM-EIA)

ACKNOWLEDGMENT
We appreciate Professor Julian Gross, Emeritus Professor of Biochemistry, Oxford University, U.K., for his careful editing and grammatical correction of this manuscript.
References
2. Yeo SI, Wong B. Current status of non-culture methods for diagnosis of invasive fungal infections. Clin Microbiol Rev. 2002. 15:465–484.

3. Stevens DA. Diagnosis of fungal infections: current status. J Antimicrob Chemother. 2002. 49:Suppl 1. 11–19.

4. Verweij PE, Poulain D, Obayashi T, Patterson TF, Denning DW, Ponton J. Current trends in the detection of antigenaemia, metabolites, and cell wall markers for the diagnosis and therapeutic monitoring of fungal infections. Med Mycol. 1998. 36:Suppl 1. 146–155.
5. Stynen D, Goris A, Sarfati J, Latge JP. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J Clin Microbiol. 1995. 33:497–500.

6. Boutboul F, Albert C, Leblanc T, Sulahian A, Gluckman E, Derouin F, Ribaud P. Invasive aspergillosis in allogeneic stem cell transplant recipients: increasing antigenemia is associated with progressive disease. Clin Infect Dis. 2002. 34:939–943.

7. Ascioglu S, Rex JH, de Pauw B, Bennett JE, Bille J, Crokaert F, Denning DW, Donnelly JP, Edwards JE, Erjavec Z, Fiere D, Lortholary O, Maertens J, Meis JF, Patterson TF, Ritter J, Selleslag D, Shah PM, Stevens DA, Walsh TJ. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin Infect Dis. 2002. 34:7–14.

9. Yoo JH, Choi JH, Choi SM, Lee DG, Shin WS, Min WS, Kim CC. Application of nucleic acid sequence-based amplification for diagnosis of and monitoring the clinical course of invasive aspergillosis in patients with hematologic diseases. Clin Infect Dis. 2005. 40:392–398.

10. Leone G, van Schijndel H, van Gemen B, Kramer FR, Schoen CD. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 1998. 26:2150–2155.

11. McClernon DR, Vavro C, St Clair M. Evaluation of a real-time nucleic acid sequence-based amplification assay using molecular beacons for detection of human immunodeficiency virus type 1. J Clin Microbiol. 2006. 44:2280–2282.

12. Ginocchio GC, Kemper M, Stellrecht KA, Witt DJ. Multicenter evaluation of the performance characteristics of the NucliSens HIV-1 QT assay used for quantitation of human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2003. 41:164–173.

13. Yates S, Penning M, Goudsmit J, Frantzen I, van de Weijer B, van Strijp D, van Gemen B. Quantitative detection of hepatitis B virus DNA by real-time nucleic acid sequence-based amplification with molecular beacon detection. J Clin Microbiol. 2001. 39:3656–3665.

14. Moore C, Hibbitts S, Owen N, Corden SA, Harrison G, Fox J, Gelder C, Westmoreland D. Development and evaluation of real-time nucleic acid sequence-based amplification assay for rapid detection of influenza A. J Med Virol. 2004. 74:619–628.
15. In : Ginocchio CC, Sillekens P, Loeffler J, van Aarie P, editors. Real-time detection of Aspergillus spp. Using nucleic acid sequence based amplification (NASBA) and molecular beacons. 2002. 12th ECCMID; Milan, Italy. –poster 1454.
16. Loeffler J, Hebart H, Cox P, Flues N, Schumacher U, Einsele H. Nucleic acid sequence-based amplification of Aspergillus RNA in blood samples. J Clin Microbiol. 2001. 39:1626–1629.
17. Hibbits S, Rahman A, John R, Westmoreland D, Fox JD. Development and evaluation of NucliSens basic kit NASBA for diagnosis of parainfluenza virus infection with 'end-point' and 'real-time' detection. J Virol Methods. 2003. 108:145–155.
18. Pinel C, Fricker-Hidalgo H, Lebeau B, Garban F, Hamidfar R, Ambroise-Thomas P, Grillot R. Detection of circulating Aspergillus fumigatus galactomannan: value and limits of the Platelia test for diagnosing invasive aspergillosis. J Clin Microbiol. 2003. 41:2184–2186.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download