Abstract
Ornithine transcarbamylase (OTC) deficiency is an X-linked co-dominant disorder. A couple, with a previous history of a neonatal death and a therapeutical termination due to OTC deficiency, was referred to our center for preimplantation genetic diagnosis (PGD). The female partner has a nonsense mutation in the exon 9 of the OTC gene (R320X). We carried out nested polymerase chain reaction (PCR) for R320X mutation and fluorescence in situ hybridization (FISH) for aneuploidy screening. Among a total of 11 embryos, two blastomeres per embryo from 9 embryos were biopsied and analyzed by duplex-nested PCR and FISH, and one blastomere per embryo from 2 embryos by only duplex-nested PCR. As a result of PCR and restriction fragment length polymorphism analysis, four embryos were diagnosed as unaffected embryos having the normal OTC gene. Among these embryos, only one embryo was confirmed as euploidy for chromosome X, Y and 18 by FISH analysis. A single normal embryo was transferred to the mother, yielding an unaffected pregnancy and birth of a healthy boy. Based on our results, PCR for mutation loci and FISH for aneuploidy screening with two blastomeres from an embryo could provide higher accuracy for the selection of genetically and chromosomally normal embryos in the PGD for single gene defects.
In the last decade, preimplantation genetic diagnosis (PGD) has become an important alternative to prenatal diagnosis for couples at high risk of transmitting inherited disorders to their children. Fluorescence in situ hybridization (FISH) has been offered for gender selection in X-linked diseases (1, 2), chromosomal translocations (3-5), aneuploidy, and recurrent implantation failure (6, 7). Procedures using polymerase chain reaction (PCR) has been applied for single gene defects including X-linked diseases (8-11).
Ornithine transcarbamylase (OTC; EC 2.1.3.3) is placed in the mitochondrial matrix where it contributes to the detoxification of nitrogenous wastes through the urea cycle, and catalyzes the synthesis of citruline from carbamyl phosphate and ornithine. The OTC gene is located on the short arm of the X chromosome within band Xp21.1 (12). It spans 73 kb, has an open reading frame of 1,062 nucleotides, and contains 10 exons and 9 introns (13, 14). OTC deficiency (MIM-#311250) is an X-linked and co-dominant metabolic disorder with partial penetrance in females (15). The phenotype of OTC deficiency is extremely heterogeneous, ranging from asymptomatic adult hemizygous males to acute neonatal hyperammonemic coma in the first week of affected male babies. Approximately 80% of heterozygous females are asymptomatic and remaining 20% show clinical severity similar to males with partial deficiencies (16, 17). There are more than 341 mutations known to cause OTC deficiency, all of which are specific for the individual families. Mutations have been found in all exons and introns with a relative paucity of mutations in the sequence encoding the leader peptide (exon 1 and a part of exon 2) and in exon 7 (17, 18).
To our knowledge, only two cases have been reported where specific PGD for OTC deficiency were carried out using multiplex or duplex-nested PCR assay and healthy babies devoid of the OTC mutation were born (19, 20). In this study, we describe the successful pregnancy and birth after PGD for OTC deficiency with simultaneous analyses of FISH and duplex-nested PCR in Korea. We have applied a duplex-nested PCR for the amplification of both the causative mutation and the Y chromosome specific Sry gene, and FISH for aneuploidy screening of chromosome X, Y and 18. A healthy boy was born by the transfer of only one unaffected normal embryo.
A couple was referred to our center after therapeutic termination of the second pregnancy following prenatal diagnosis for OTC deficiency. In the first pregnancy, a male baby was born but died at 10 days after birth due to liver dysfunction accompanying hyperammonemia. Molecular genetic analysis of the couple's OTC gene was performed by PCR and direct sequencing at Seoul Asan Hospital and revealed a single base substitution (R320X) in the exon 9 of the female partner. In the second pregnancy, the fetus was identified to be an affected boy by PCR and direct sequencing on a chorionic villus sample, and the pregnancy was therapeutically terminated. In order to avoid a pregnancy with an affected baby, the couple was counseled for PGD in Cheil General Hospital.
In order to determine the efficiency of single cell PCR and allele drop-out (ADO) rate, primers used for the detection of the causative mutation and Y chromosome specific Sry gene were tested using single lymphocytes (Table 1). Ovarian stimulation, in vitro fertilization, and blastomere biopsy were done as previously described (4, 5). After the blastomere biopsy procedure, the embryos were washed several times and transferred to the G2.2 medium (Vitrolife Sweden AB, Kungsbacka, Sweden). Each blastomere was washed twice through two drops of G2.2 medium and transferred into sterile 0.2 mL PCR tubes containing 5 µL of alkaline lysis buffer. For each embryo biopsied, blank negative controls were prepared from the wash drops. The nested-duplex PCR analysis was performed by previously described protocol (6). Mutation analysis by restriction fragment length polynorphism (RFLP) with BclI restriction enzyme was carried out only for positively amplified PCR products of blastomeres. Another blastomere from the same embryo was prepared for FISH. The FISH analysis for chromosome X, Y and 18 was done as previously described and the manufacture's protocol (4, 5). The embryos were cultured under standard culture condition until the diagnosis was established. The embryos with the normal genotype were selected and transferred on day 4 after fertilization. Amniocentesis was performed at 16 weeks of gestation to confirm the results of PGD. Postnatal genetic and physiological diagnosis was also conducted.
All 10 exons and exon-intron boundaries of the OTC gene were screened by PCR followed by direct DNA sequencing analysis. The heterozygous OTC gene with R320X nonsense mutation (CGA→TGA) and wild type was detected in the exon 9 of the female partner by RFLP (Fig. 1) and direct sequencing (data not shown). This R320X nonsense mutation was associated with acute neonatal hyperammonemia in their first pregnancy and reduced enzyme activity by 10-15% (21). Male partner has a normal OTC gene.
The efficiency and accuracy of the duplex-nested PCR analysis were tested on single lymphocytes from the heterozygous female partner and normal male partner. PCR on single lymphocytes resulted in 90.6% (58/64) amplification rate, whereas none of the negative controls showed a positive band. ADO was detected in 4 out of 30 lymphocytes (13.3%).
After ovarian hyperstimulation, a total of 18 oocytes were retrieved and 17 oocytes were injected using intracytoplasmic sperm injection in order to prevent DNA contamination from inseminated sperm. Among a total of 11 embryos, two blastomeres per embryo from 9 embryos were biopsied and simultaneously analyzed by duplex-nested PCR and FISH, and one blastomere per embryo from 2 embryos by only duplex-nested PCR. As a result of duplex-nested PCR and RFLP analysis with the BclI restriction enzyme, ten showed successful amplification for the exon 9 of the OTC gene, and the amplification rate was 90.9%, almost same as that of the single lymphocyte test. There were two unaffected males, five affected males, two unaffected females, and one heterozygous female (Fig. 2 and Table 2). No amplification was detected in any of the negative controls. On the other hand, out of nine blastomeres analyzed by FISH, only six blastomeres were identified as chromosomally normal embryos (Fig. 3, Table 2). However, among total four unaffected embryos (embryo 1, 2, 3, and 8) diagnosed by PCR, only two embryos (embryo 3 and 8) were identified as chromosomally normal by FISH. The other two embryos (embryo 1 and 2) were shown to have trisomy 18 and monosomy 18, respectively. Only one male embryo (embryo 8) was transferred, and another female unaffected embryo (embryo 3) was cryopreserved at blastocyst stage on day 5 after fertilization. This PGD cycle resulted in a singleton pregnancy and the genotype of the fetus was confirmed to be a normal male by amniocentesis at 16 weeks of gestation. A healthy boy of 2.8 kg was delivered at 36 weeks. The genotype of the baby for the exon 9 of OTC gene and level of ammonia were confirmed as normal.
Most PGD cycles for X-linked disorders such as Duchenne muscular dystrophy and fragile X syndrome involved the selection of female embryos which were diagnosed as normal or heterozygous. These gender selections were supported by couples at risk of X-linked recessive conditions. However, OTC deficiency is an X-linked and co-dominant disorder. Some heterozygous females represent severe phenotypic abnormalities such as nausea, vomiting and ataxia. ADO for affected allele could lead to yielding an affected heterozygous baby, even if the female embryo was diagnosed as normal by PGD for OTC deficiency. Gender selection with FISH for Y chromosome-specific loci is more reliable because amplification failure could occasionally occur in single cell PCR. Thus, our protocol with PCR and FISH in this study provides higher accuracy for the selection of genetically and chromosomally normal embryos in the PGD for X-linked co-dominant defects.
We applied duplex-nested PCR and FISH analysis in two blastomeres from each embryo, which were more than 6-cell stage. Biopsy of more than a quarter of blastomeres from each embryo could interfere with developmental potential of embryos. Thus, single blastomere biopsy was applied to less than 6-cell stage embryos in this study. Sequential PCR and FISH analysis in one blastomere (cell recycling) was reported and could be used to detect the aneuploidy, but the rate of ADO after PCR seems to be higher using this method (22, 23). Therefore, we carried out two methods separately on two blastomeres. A duplex-nested PCR analysis was developed for the simultaneous diagnosis of a R320X mutation and a gender selection, and FISH analysis was used to detect aneuploidy for chromosome X, Y and 18. As a result of duplex nested PCR and FISH analysis, concordant results have been obtained from two blastomeres at the sex locus, and the amplification efficiency was 100% (9/9 except two blastomeres, that were not analyzed by FISH analysis) for Sry. And the amplification rate was 90.9% (10/11) for exon 9 of the OTC gene.
Several strategies have been developed to decrease amplification failure and ADO in single cell analysis of PGD. These included the development of sequential first and second polar body analysis (24), improvement of the PCR condition by increasing the denaturation temperature and time (19), using a more powerful lysis method (25-28) and the use of fluorescent PCR for mutations or linked markers and multiplex PCR (29-32). As mentioned above, we have also endeavored to optimize single cell PCR conditions, which included longer initial denaturation time, higher denaturation temperature, and the selection of Taq polymerase, the concentration of MgCl2, and lysis methods. The incidence of ADO was 5-15% in many PGD laboratories (27, 28). At a preliminary experiment on single lymphocytes in this study, the ADO rate for exon 9 of the OTC gene was about 13.0% when using duplex-nested PCR. We could not absolutely exclude the possibility of ADO in the diagnosis of unaffected female embryo owing to the relatively higher ADO. Thus, FISH was simultaneously applied for the aneuploidy screening and sex selection to prevent the pregnancy of heterozygous female.
In this study, we report the successful PGD and delivery of a healthy boy in a couple at risk of transmitting the OTC deficiency. This was carried out by the simultaneous analysis of the duplex-nested PCR for a R320X mutation in the OTC gene underlying the OTC deficiency and Y chromosome-specific loci, and FISH for aneuploidy screening of chromosome X, Y and 18. This strategy could provide higher accuracy for the selection of genetically and chromosomally normal embryos in PGD for single gene defects.
Figures and Tables
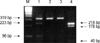 | Fig. 1Identification of the maternal mutation (heterozygote type) in exon 9 of the OTC gene. After digesting the PCR product with BclI, the 319 or 218 bp PCR products of the affected mother were digested into 223 and 96 bp (lane 1) and 178 and 40 bp products (lane 4), respectively. The 40 bp product was hardly seen on an agarose gel. Lane 1 and 4; the first and nested PCR products of the affected mother, respectively, lane 2 and 3; BclI-digested products from the first PCR products of the father. M: molecular weight marker. |
 | Fig. 2Results of agarose gel electrophoresis of the restriction fragment length polymorphism (RFLP) analysis with BclI restriction enzyme in the preimplantation genetic diagnosis for ornithine transcarbamylase deficiency. In normal embryos (No. 1, 2, 3 and 8), the 218-bp PCR products were not digested with BclI. |
 | Fig. 3Results of the fluorescence in situ hybridization (FISH) analysis on the embryos diagnosed to be normal by PCR in the PGD for OTC deficiency. Panels A-D show the photographs for the results of FISH on the embryos No. 1, 2, 3, and 8 in Table II, respectively. Aqua, green and orange signals indicate the presence of the chromosome X, Y and 18 in the nucleus of the blastomeres, respectively.
PGD, preimplantation genetic diagnosis; OTC, ornithine transcarbamylase; PCR, polymerase chain reaction
|
Table 1
Sequences of oligonucleotide primers and PCR conditions for duplex-nested PCR for OTC deficiency

Table 2
Summary of the results of the PGD for OTC deficiency with simultaneous analysis of duplex nested PCR and FISH

Plus (+) and minus (-) signs indicate whether a band was detected corresponding to the Sry gene. N.A. (not assayed) indicates that blastomere was not available for FISH. In the table, transferred embryo is presented with bold and italic characters.
PGD, preimplantation genetic diagnosis; OTC, ornithine transcarbamylase; PCR, polymerase chain reaction; FISH, fluorescence in situ hybridization.
References
1. Munne S, Sultan KM, Weier HU, Grifo JA, Cohen J, Rosenwaks Z. Assessment of numerical abnormalities of X, Y, 18 and 16 chromosomes in preimplantation embryos before transfer. Am J Obstet Gynecol. 1995. 172:1191–1201.
3. Delhanty JD. Chromosome analysis by FISH in human preimplantation genetics. Hum Reprod. 1997. 12(11):Suppl. 153–155.
4. Lim CK, Jun JH, Min DM, Lee HS, Kim JY, Koong MK, Kang IS. Efficacy and clinical outcome of preimplantation genetic diagnosis using FISH for couples of reciprocal and Robertsonian translocations: the Korean experience. Prenat Diagn. 2004. 24:556–561.
5. Lim CK, Min DM, Lee HS, Byun HK, Park SY, Ryu HM, Kim JY, Koong MK, Kang IS, Jun JH. Improvement of pregnancy rate in preimplantation genetic diagnosis with FISH procedure by the laboratory optimization and experiences. Korean J Fertil Steril. 2004. 31:29–39.
6. Gianaroli L, Magli MC, Ferraretti AP, Munne S. Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: identification of the categories for which it should be proposed. Fertil Steril. 1999. 72:837–844.

7. Munne S, Magli C, Cohen J, Morton P, Sadowy S, Gianaroli L, Tucker M, Marquez C, Sable D, Ferraretti AP, Massey JB, Scott R. Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod. 1999. 14:2191–2199.
8. Lissens W, Sermon K. Preimplantation genetic diagnosis: current status and new development. Hum Reprod. 1997. 12:1756–1761.
9. Ray PF, Handyside AH. Increasing the denaturation temperature during the first cycles of amplification reduces allele dropout from single cells for preimplantation genetic diagnosis. Mol Hum Reprod. 1996. 2:213–218.
10. Malcov M, Schwartz T, Mei-Raz N, Yosef DB, Amit A, Lessing JB, Shomrat R, Orr-Urtreger A, Yaron Y. Multiplex nested PCR for preimplantation genetic diagnosis of spinal muscular atrophy. Fetal Diagn Ther. 2004. 19:199–206.

11. Lee HS, Choi HW, Lim CK, Koong MK, Kang IS, Yoo HW, Choi JH, Jun JH. Identification of a novel single nucleotide polymorphism of HADHA gene at a referred primer-binding site during pre-diagnostic tests for preimplantation genetic diagnosis. J Korean Med Sci. 2006. 21:794–799.

12. Lindgren V, de Martinville B, Horwich AL, Rosenberg LE, Francke U. Human ornithine transcarbamylase locus mapped to band Xp21.1 near the Duchenne muscular dystrophy locus. Science. 1984. 226:698–700.

13. Horwich AL, Fenton WA, William KR, Kalousek F, Kraus JP, Doolittle RF, Konigsberg W, Rogenberg LE. Structure and expression of a complementary DNA for the nuclear coded precursor of human mitochondrial ornithine transcarbamylase. Science. 1984. 224:1068–1074.

14. Hata A, Tsuzuki T, Shimada K, Takiguchi M, Mori M, Matsuda I. Structure of the human ornithine transcarbamylase deficiency. Am J Hum Genet. 1988. 48:212–222.
15. Pelet A, Rotig A, Bonaiti-Pellie C, Rabier D, Cormier V, Toumas E, Hentzen D, Saudubray JM, Munnich A. Carrier detection in a partially dominant X-linked disease: ornithine transcarbamylase deficiency. Hum Genet. 1990. 84:167–171.

16. Hudak ML, Douglas Jones M, Brusilow SW. Differentiation of transient hyperammonemia of the newborn and urea cycle enzyme defects by clinical presentation. J Pediatr. 1985. 107:712–719.

17. Tuchman M, Jaleel N, Morizono H, Sheehy L, Lynch MG. Mutations and polymorphisms in the human ornithine transcarbamylase gene. Hum Mutat. 2002. 19:93–107.

18. Yamaguchi S, Brailey LL, Morizono H, Bale AE, Tuchman M. Mutation and polymorphisms in the human ornithine transcarbamylase (OTC) gene. Hum Mutat. 2006. 27:626–632.
19. Ray PF, Gigarel N, Bonnefont JP, Attie T, Hamamah S, Frydman N, Vekemans M, Frydman R, Munnich A. First specific preimplantation genetic diagnosis for ornithine transcarbamylase deficiency. Prenat Diagn. 2000. 20:1048–1054.

20. Verlinsky Y, Rechitsky S, Verlinsky O, Strom C, Kuliev A. Preimplantation diagnosis for ornithine transcarbamylase deficiency. Reprod Biomed Online. 2000. 1:45–47.

21. Yoo HW, Kim GH, Lee DH. Identification of new mutations in the ornithine transcarbamylase (OTC) gene in Korean families. J Inherit Metab Dis. 1996. 19:31–42.

22. Rechitsky S, Freidine M, Verlinsky Y, Strom CM. Allele dropout in sequential PCR and FISH analysis of single cells (cell recycling). J Assist Reprod Genet. 1996. 13:115–124.

23. Thornhill AR, Monk M. Cell recycling of a single human cell for preimplantation diagnosis of X-linked disease and dual sex determination. Mol Hum Reprod. 1996. 2:285–289.
24. Strom CM, Ginsberg N, Rechitsky S, Cieslak J, Ivakhenko V, Wolf G, Lifchez A, Moise J, Valle J, Kaplan B, White M, Barton J, Kuliev A, Verlinsky Y. Three births after preimplantation genetic diagnosis for cystic fibrosis with sequential first and second polar body analysis. Am J Obstet Gynecol. 1998. 178:1298–1306.

25. Wu K, Cuppens H, Buyse I, Decorte R, Marynen P, Gordts S, Cassiman JJ. Co-amplification of the cystic fibrosis delta F508 mutation with the HLA DQA1 sequence in single cell PCR: implications for improved assessment of polar bodies and blastomeres in preimplantation diagnosis. Prenat Diagn. 1993. 13:1111–1112.
26. Thornhill AR, McGrath JA, Eady RA, Braude PR, Handyside AH. A comparison of different lysis buffers to assess allele dropout from single cells for preimplantation genetic diagnosis. Prenat Diagn. 2001. 21:490–497.

27. Piyamongkol W, Bermudez MG, Harper JC, Wells D. Detailed investigation of factors influencing amplification efficiency and allele dropout in single cell PCR: implications for preimplantation genetic diagnosis. Mol Hum Reprod. 2003. 9:411–420.

28. Choi HW, Lee HS, Lim CK, Koong MK, Kang IS, Jun JH. Reliability of the single cell PCR analysis for preimplantation genetic diagnosis of single gene disorders. Korean J Fertil Steril. 2005. 32:293–300.
29. Dreesen J, Jacobs L, Bras M, Herberg J, Dumoulin JC, Geraedts JP, Evers JL, Smeets HJ. Multiplex PCR of polymorphic markers flanking the CFTR gene: a general approach for preimplantation genetic diagnosis of cystic fibrosis. Mol Hum Reprod. 2000. 6:881–885.

30. Eftedal I, Schwartz M, Bendtsen H, Andersen A, Zieve S. Single intragenic microsatellite preimplantation enetic diagnosis for cystic fibrosis provides positive allele identification of all CFTR genotypes for informative couples. Mol Hum Reprod. 2001. 7:307–312.
31. Goossens V, Sermon K, Lissens W, Vandervorst M, Vanderfaeillie A, De Rijcke M, De Vos A, Henderix P, Van De Velde H, Van Steirteghem A, Liebaers I. Clinical application of preimplantation genetic diagnosis for cystic fibrosis. Prenat Diag. 2000. 20:571–581.

32. Lee HS, Choi HW, Lim CK, Park SY, Kim JY, Koong MK, Jun JH, Kang IS. Efficacy of duplex-nested PCR and fluorescent PCR in the preimplantation genetic diagnosis for Duchenne muscular dystrophy. Korean J Fertil Steril. 2005. 32:17–26.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download