Abstract
Recovery from hepatitis B virus (HBV) infection depends on the cellular immune responses. Chemokines and their receptors play significant roles in immune defense. This study was undertaken to investigate the association between HBV infection and single nucleotide polymorphisms (SNPs) of genes for the chemokines and their receptors. Between March 2002 and February 2004, a total of 957 single ethnic Korean patients were enrolled into two different groups; "HBV clearance group" (n=350), who have recovered from HBV infection, and "HBV persistence group" (n=607), who were repeatedly HBsAg-positive. The HBV persistence group was subdivided into "inactive carrier" and "HBV progression group (chronic hepatitis and cirrhosis)". We assessed polymorphisms in regulated and normal T-cell expressed and secreted (RANTES) at position -403, monocyte chemoattractant protein-1 (MCP-1) at position -2518, CCR2 V64I, CCR5 -2459, CXCR1 S276T and CXCR4 I138I using single primer extension assay. Genotype distributions of the "HBV clearance versus persistence group" and "inactive carrier versus HBV progression group" were compared. On the basis of unconditional logistic regression analysis with adjustment for age and sex, no statistically significant association with susceptibility to persistent HBV infection was observed with RANTES -403, MCP-1 -2518, CCR2 V64I, CCR5 -2459, CXCR1 S276T, and CXCR4 I138I polymorphisms. In addition, no association of analyzed SNPs with HBV disease progression was found.
Hepatitis B virus (HBV) infection is one of the most important chronic liver diseases worldwide, especially in several areas of Asia and Africa. More than 350 million people in the world are persistently infected with HBV and are at risk of developing chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). Age at infection is a well-known determinant of progression to chronicity (1-3). In adults with normal immunological status, less than five percent of individuals with an acute hepatitis B infection go on to develop a chronic infection, characterized by persistence of circulating viral antigens and viral DNA. On the other hand, the rate of evolution to the chronic state is much higher when the infection is acquired in childhood, although the initial infection is usually clinically asymptomatic. However, the underlying mechanisms for the persistence of HBV or HCC occurrence are not fully understood.
There is growing evidence that genetic variation plays an important role in the determination of individual susceptibility to complex disease traits. It has been thought that genetic associations may also provide a clue to the outcome to HBV infection. Several lines of evidence suggest that genetic polymorphisms are involved in susceptibility to persistent HBV infection or development of HCC (4-11).
Chemokines are a groups of chemotactic molecules that specifically attract and recruit populations of immune effector cells to the sites of injury or infection (12). Their major role is in leukocyte migration and dependent processes such as immune surveillance and innate and adaptive immune responses. Apart from T-cell migration, CCR5 and other chemokine receptors mediate cell activation, costimulation, and differentiation of T-cells and monocyte during innate and adaptive immune response (13). The production of some chemokines varies among individuals, and these variations may be determined by genetic polymorphisms.
Regulated and normal T-cell expressed and secreted (RANTES) may have a role in the selective recruitment of T-cells to portal and periportal regions in liver during inflammation (14). It might be expected that the less frequent allele in RANTES gene would be associated with increased portal inflammation, interface hepatitis, and severity of fibrosis. Two single nucleotide polymorphisms (SNPs) have been described in the RANTES promoter region, including the G-to-A substitution at position -403. There is evidence that the mutant A allele leads to increased transcription of RANTES (15).
Monocyte chemoattractant protein-1 (MCP-1) is active on multiple leukocyte populations showing chemotactic activity at a low concentration in vitro (16). Hepatic stellate cells (HSCs) have been shown to regulate leukocyte trafficking by secreting MCP-1, and it has been suggested that MCP-1 may have a direct profibrogenic action via HSC chemotaxis (17). In vitro cells from individuals who are heterozygous or homozygous for G at -2518 appear to produce more MCP-1 than cells from individuals homozygous for A at -2518 (18). Hepatitis C virus (HCV) infected patients with the G allele at position -2518 in the MCP-1 gene were reported to be more prone to hepatic inflammation and fibrosis (19).
CCR5 was shown to act as a cofactor for entry of macrophage-tropic strains of HIV-1 (20). Moreover, CCR5 is a candidate gene for the outcome of hepatitis C virus infection. It was reported that the frequency of the HIV-protective CCR5-Δ32/Δ32 genotype is increased in hepatitis C (21).
CCR2 has a role in modulating the immune response as well as recruiting monocyes/macrophages to the sites of inflammation has been suggested (22). One variant has been reported in CCR2, which leads to a valine-to-isoleucine substitution at amino acid position 64 (23). CCR2 V64I SNP was reported to be associated with a delay in progression to AIDS, and CCR2 was thought to the one of HIV-1 co-receptor (24).
CXCR1 (interleukin-8 receptor alpha) is a chemokine receptor, which is known to have IL-8 as a binding ligand (25). CXCR4 function as a coreceptor for T-cell line trophic strains of HIV-1 (26).
Although chemokines and their receptors have received increased attention, the role of chemokines and their receptors in HBV infection has not been clarified. A series of studies using transgenic mouse models have demonstrated that CD8+ cells have the capacity to non-cytologically clear HBV from hepatocytes that replicate HBV encoded by a transgene (27). The protective role of HBV-specific CD8+ cells is dependent on their ability to efficiently migrate to the infected liver, where they may exert an effector function. The migratory behavior of CD8+ cells is influenced by their expression of different chemokines or their receptors. A recent report showed that the ability of circulating HBV-specific CD8+ cells of patients with low replicating virus to upregulate CCR5 suggests that these cells may respond to increases in virus replication by efficiently migrating into the infected liver (28).
Therefore, we hypothesized that several important chemokines and their receptors are deemed to be related to the outcome of HBV infection. The purpose of this study is to ascertain whether the genetic polymorphisms of RANTES, MCP-1, CCR2, CCR5, CXCR1, and CXCR4 genes in HBV infection could influence their outcomes in terms of viral clearance or disease progression.
Between March 2002 and February 2004, a total of 957 patients were included in this study, and they were enrolled from outpatient clinic of the Gastroenterology Department and from the Center for Health Promotion of Ajou University Hospital, Suwon, Korea. All the patients were single ethnic Korean subjects. They were classified into three groups according to the various status of HBV infection; "HBV Clearance group" (7) [n=350, HBsAg (-), AntiHBc (+), AntiHBs (+)], "Inactive Carrier group" [n=149, HBsAg (+) & HBeAg (-), sustained normal transaminase] and "HBV progression group (Chronic Hepatitis or Liver Cirrhosis, irrespective of HBeAg positivity)" [n=458, HBsAg (+), elevated transaminase (≥2 times the upper limit of normal) or evidence of cirrhosis]. Among them, inactive carrier and HBV progression groups (a total of 607 patients) were classified as the "HBV persistence" group who continued to be positive for HBsAg for more than 6 months. Patients who were positive for anti-HBs without anti-HBc, and patients with other types of chronic liver disease were excluded from the analysis. Informed consent was obtained from each patient, and the Institutional Review Board of Human Research of Ajou University Hospital approved the study protocol.
To elucidate their role in the immunopathogenesis of HBV infection, six polymorphisms were studied in two chemokine (RANTES -403, MCP-1 -2518) and four chemokine receptor (CCR2 V64I, CCR5 -2459, CXCR1 S276T, CXCR4 I138I) genes in Korean patients with HBV infection, and in HBsAg negative controls who had eliminated HBV.
Genomic DNA was extracted from 300 µL whole blood using a DNA Purification Kit (GENTRA, Minneapolis, MN, U.S.A.) according to the manufacturer's instructions. Six target polymorphisms were detected by polymerase chain reaction (PCR) amplification.
The sequence of the primers and probes used in the assays are provided in Table 1. The parameters for thermocycling were as follows: an initial activation step of 95℃ for 10 min preceded the cycling program. All amplification conditions were 35 cycles of 30 sec at 95℃ for denaturation, 1 min at 60℃ for annealing, followed by a single 10-min extension cycle at 72℃. The polymorphisms were detected by single nucleotide primer extension assay (SNP IT, Beckman Coulter, Fullerton, CA, U.S.A.) using the method as previously described (15). Briefly, the genomic DNA region spanning the polymorphic site was PCR-amplified using one phosphorothiolated primer and one regular PCR primer. The amplified PCR products were digested with exonuclease. The 5'-phosphorothioates protect one strand of the PCR product from exonuclease digestion, resulting in the generation of a single-stranded PCR template. The single-stranded PCR template is overlaid onto a 384-well plate that contains covalently attached SNP-IT extension primer designed to hybridize immediately adjacent to the polymorphic site. The SNP-IT primer is extended for a single base with DNA polymerase and mixture of appropriate acycloterminator, which is labeled with either fluorescein isothiocyanate (FITC) or biotin and is complementary to the polymorphic nucleotide. The identity of the incorporated nucleotide is determined with serial colorimetric reactions with anti-FITC-alkaline phosphatase and streptavidin-horseradish peroxidase, respectively. The results of yellow and/or blue color developments were analyzed with ELISA reader and the final genotype calls were made with the QCReview program (Beckman Coulter, Fullerton, CA, U.S.A.).
Hardy-Weinberg equilibrium of alleles at individual loci was assessed by χ2-statistics. Demographic characteristics of patients with HBV persistence or HBV clearance were compared using χ2-tests on categorical variables and independent sample t-tests on normally distributed continuous variables. Multivariate analysis was performed after adjusting for age and sex as co-variables using SPSS version 11.0 software (SPSS, Chicago, IL, U.S.A.). For multivariate analysis, binary logistic regression analysis was performed to determine which factor(s) was the most discriminating for HBV persistence or the disease progression in chronic hepatitis B infection, where age, sex, and SNPs of each gene were independent variables. Selection of variables was done by backward stepwise deletion. All p values were two-tailed, and p value <0.05 were considered to indicate statistical significance.
Among six SNPs tested in this study, three SNPs were located in the regulatory regions (RANTES -403, MCP-1 -2518, CCR5 -2459) and three were in the coding regions (CCR2 V64I, CXCR1 S276T and CXCR4 I138I) of genes. In case of CXCR1 and CXCR4 genes, one non-synonymous SNP at codon 276 in CXCR1 coding sequences (S276T) and one silent SNP at codon 138 in CXCR4 coding sequences (I138I) were detected in our study population. Other sites were well-known SNPs, which had been frequently reported.
Genotype distributions at all polymorphic sites approximated the Hardy-Weinberg equilibrium (Table 2). Allelic variations were compared among HBV clearance and persistence group and in the persistence group, inactive healthy carrier, and the disease progression group including chronic hepatitis and cirrhosis.
Demographic characteristics of the subjects with HBV clearance and persistence groups were shown in Table 3. Differences between two groups in aspartate transaminase/alanine transaminase (AST/ALT), bilirubin, alpha-fetoprotein (AFP) were observed. The serum levels of AST, ALT, bilirubin, and AFP at baseline were significantly higher in the HBV persistence group. No significant differences were detected between the clearance group and the persistence group in the distribution of gender, but the age tended to be younger in the persistence group.
All SNP frequencies were analyzed for an association with spontaneous clearance versus HBV persistence. Genotype distributions and p-value for logistic analysis whilst controlling for age and sex as covariates are shown (Table 4). In multivariate analysis after adjustment for age and sex, there was no significant difference in the frequencies of allele in the analyzed genes between the HBV clearance group and the HBV persistence group (Table 4).
For the further analysis of the relationship between chemokine/chemokine receptor gene SNPs and disease progression in HBV infection, the persistence group was divided into non-progressive group (inactive carrier) and the progression group (chronic hepatitis and liver cirrhosis).
Demographic characteristics of the subjects with "inactive carrier" vs. "HBV progression" group are summarized in Table 5. No significant differences were identified between two groups in the distributions of their age, but male gender was predominant in the disease progression group. AST, ALT, albumin, platelets were different between the two groups. The serum levels of AST, ALT at baseline were significantly higher in HBV progression group.
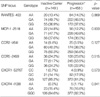
Table 6 demonstrates the allele frequencies of polymorphic loci investigated between the inactive carrier and HBV progression groups. No statistically significant relationship was seen between genetic polymorphisms in chemokines and their receptor genes and the HBV disease progression.
This is a large case-control study, looking at six SNPs in a group of chemokines and their receptors in single ethnic population. Six polymorphisms in chemokine-related genes were analyzed in 607 Korean HBV carriers and 350 who had eliminated HBV as a control. The allelic frequencies of investigated genes in "HBV clearance group" did not differ from the frequencies in "HBV persistence group". No significant differences in the distribution of genotypes associated with chemokines and their receptor gene polymorphisms could be demonstrated between "Inactive carrier group" and "HBV progressive group" in this study.
In Korea, vertical transmission of HBV in infancy is a major cause of chronic infection. The age at which an individual is infected determines the likelihood of the development of chronic infection. Thus, ethnic differences in the mode of infection and the distribution of allelic polymorphisms may account for the lack of the correlation in our study. Common methodologic limitations of genetic epidemiologic studies include a low sample size leading to a lack of statistical power. In the present study, we analyzed a large number of cases (n=957) of a single ethnic origin, therefore our results may raise the statistical power of this study.
During the last few years, numerous association studies have investigated the role of genetic polymorphisms in the outcome of HBV infection (4-9). We also have shown that the interleukin-10 and tumor necrosis factor-α promoter SNPs influenced the HBV clearance after acute infection (11). Chemokines and their receptors have received increasing attention in the last few years. It is now clear that chemokines participate in many pathological conditions like inflammation and are likely key regulators of immune responses (25). However, there are few reports about association between chemokine related gene SNPs and the outcome of HBV infection.
This study has some limitations. The natural course of chronic hepatitis B can be divided into several phases based on virus-host interaction; immune tolerance phase, immune clearance phase, inactive carrier or chronic hepatitis, eventually cirrhosis, and/or hepatocelluar carcinoma. In this study, the family history of HBV infection could not be completely evaluated, and it was hard to presume the onset of HBV infection. Moreover, inactive carrier and progression groups could not be exactly divided because most enrolled subjects did not undergo liver biopsy. Even though we tried to compensate the confounding effects by adjusting age and sex, limitations from a cross-sectional study could not be neglected. Unfortunately, we could not categorize the patient population by virological status (HBeAg and HBV-DNA) due to incomplete data.
CCR5 is a strong candidate gene for the outcome of HCV infection.(21) But their roles in HBV infection are not clear. In a recent investigation, Chang et al. have shown that CCR5 promoter SNPs might influence the HBV clearance in Korean population (29), whereas our study did not find any such correlation in the same ethnic Korean population. Discrepant results may result from demographic factors and differences in the study design.
Recently, MCP-1 gene SNP was reported to be associated with hepatic MCP-1 expression and the severity of HCV-related liver disease (19). Although there is no evidence of a role for MCP-1 in the pathogenesis of HBV in the literature, it is a potential candidate gene because it could affect the susceptibility to persistent HBV infection, inflammatory response, or liver fibrosis. The results from this study showed no significant association with susceptibility to persistent HBV carriage. Such discrepancy may reflect differences in the immunopathogenesis of HBV and HCV infection.
Although none of the SNPs studied are likely to have major effects on the outcome of HBV infection, we should consider that chemokine expression may be disease stage-specific, and serum levels may not necessarily correlate with hepatic expression. Determination of the exact functional consequences of these polymorphisms need more detailed and careful in vitro as well as in vivo studies.
In conclusion, this study has attempted to elucidate the role of genetic polymorphisms in chemokines and their receptors in the outcome of HBV infection, but showed no association between HBV clearance or disease progression in HBV infection and chemokines or chemokine receptors gene polymorphisms. Further studies are needed to determine whether an association exists between the outcome of HBV infection including antiviral treatment response of chronic hepatitis B and genetic polymorphisms in other chemokines.
Figures and Tables
Table 4
Logistic regression analysis of polymorphisms with the HBV clearance whilst controlling for age and sex as covariates
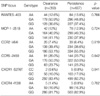
Table 6
Logistic regression analysis of polymorphisms with the HBV clearance whilst controlling for age and sex as covariates
*Chronic hepatitis or liver cirrhosis.
For multivariate analysis, binary logistic regression analysis was performed after adjusting for age and sex as co-variables. Selection of variables was done by backward stepwise deletion.
SNP, single nucleotide polymorphisms; MCP-1, monocyte chemoattractant protein-1.
References
1. Stevens CE, Beasley RP, Tsui J, Lee WC. Vertical transmission of hepatitis B antigen in Taiwan. N Engl J Med. 1975. 292:771–774.

2. Coursaget P, Yvonnet B, Chotard J, Vincelot P, Sarr M, Diouf C, Chiron JP, Diop-Mar I. Age- and sex-related study of hepatitis B virus chronic carrier state in infants from an endemic area (Senegal). J Med Virol. 1987. 22:1–5.

3. Tassopoulos NC, Papaevangelou GJ, Sjogren MH, Roumeliotou-Karayannis A, Gerin JL, Purcell RH. Natural history of acute hepatitis B surface antigen-positive hepatitis in Greek adults. Gastroenterology. 1987. 92:1844–1850.

4. Ahn SH, Han KH, Park JY, Lee CK, Kang SW, Chon CY, Kim YS, Park K, Kim DK, Moon YM. Association between hepatitis B virus infection and HLA-DR type in Korea. Hepatology. 2000. 31:1371–1373.

5. Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003. 98:144–150.

6. Hohler T, Kruger A, Gerken G, Schneider PM, Meyer zum Buschenefelde KH, Rittner C. A tumor necrosis factor-alpha (TNF-alpha) promoter polymorphism is associated with chronic hepatitis B infection. Clin Exp Immunol. 1998. 111:579–582.
7. Kim YJ, Lee HS, Yoon JH, Kim CY, Park MH, Kim LH, Park BL, Shin HD. Association of TNF-alpha promoter polymorphisms with the clearance of hepatitis B virus infection. Hum Mol Genet. 2003. 12:2541–2546.
8. Shin HD, Park BL, Kim LH, Jung JH, Kim JY, Yoon JH, Kim YJ, Lee HS. Interleukin 10 haplotype associated with increased risk of hepatocellular carcinoma. Hum Mol Genet. 2003. 12:901–906.

9. Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995. 332:1065–1069.

10. Cheong JY, Cho SW, Chung SG, Lee JA, Yeo M, Wang HJ, Lee JE, Hahm KB, Kim JH. Genetic Polymorphism of Interferon-gamma, Interferon-gamma Receptor, and Interferon Regulatory Factor-1 Genes in Patients with Hepatitis B Virus Infection. Biochem Genet. 2006. 44:246–255.
11. Cheong JY, Cho SW, Hwang IL, Yoon SK, Lee JH, Park CS, Lee JE, Hahm KB, Kim JH. Association between chronic hepatitis B virus infection and interleukin-10, tumor necrosis factor-alpha gene promoter polymorphisms. J Gastroenterol Hepatol. 2006. 21:1163–1169.

12. Borish LC, Steinke JW. 2. Cytokines and chemokines. J Allergy Clin Immunol. 2003. 111:S460–S475.

13. Strieter RM, Standiford TJ, Huffnagle GB, Colletti LM, Lukacs NW, Kunkel SL. "The good, the bad, and the ugly." The role of chemokines in models of human disease. J Immunol. 1996. 156:3583–3586.
14. Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996. 35:3362–3367.

15. An P, Nelson GW, Wang L, Donfield S, Goedert JJ, Phair J, Vlahov D, Buchbinder S, Farrar WL, Modi W, O'Brien SJ, Winkler CA. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci USA. 2002. 99:10002–10007.

16. Loetscher P, Seitz M, Clark-Lewis I, Baggiolini M, Moser B. Monocyte chemotactic proteins MCP-1, MCP-2, and MCP-3 are major attractants for human CD4+ and CD8+ T lymphocytes. Faseb J. 1994. 8:1055–1060.
17. Marra F, Romanelli RG, Giannini C, Failli P, Pastacaldi S, Arrighi MC, Pinzani M, Laffi G, Montalto P, Gentilini P. Monocyte chemotactic protein-1 as a chemoattractant for human hepatic stellate cells. Hepatology. 1999. 29:140–148.

18. Rovin BH, Lu L, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem Biophys Res Commun. 1999. 259:344–348.

19. Muhlbauer M, Bosserhoff AK, Hartmann A, Thasler WE, Weiss TS, Herfarth H, Lock G, Scholmerich J, Hellerbrand C. A novel MCP-1 gene polymorphism is associated with hepatic MCP-1 expression and severity of HCV-related liver disease. Gastroenterology. 2003. 125:1085–1093.

20. Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996. 381:667–673.

21. Woitas RP, Ahlenstiel G, Iwan A, Rockstroh JK, Brackmann HH, Kupfer B, Matz B, Offergeld R, Sauerbruch T, Spengler U. Frequency of the HIV-protective CC chemokine receptor 5-Delta32/Delta32 genotype is increased in hepatitis C. Gastroenterology. 2002. 122:1721–1728.
22. Kurihara T, Warr G, Loy J, Bravo R. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J Exp Med. 1997. 186:1757–1762.

23. Kostrikis LG, Huang Y, Moore JP, Wolinsky SM, Zhang L, Guo Y, Deutsch L, Phair J, Neumann AU, Ho DD. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998. 4:350–353.

24. Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, Goedert JJ, O'Brien TR, Jacobson LP, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner MW, O'Brien SJ. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997. 277:959–965.
25. Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000. 18:217–242.

26. Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997. 94:1925–1930.

27. Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996. 4:25–36.

28. Lee CK, Suh JH, Cho YS, Han KH, Chung JB, Chon CY, Moon YM. Chemokine receptor expression of hepatitis B virus-specific CD8+ lymphocyte in chronic B viral infection. Taehan Kan Hakhoe Chi. 2002. 8:363–370.
29. Chang HY, Ahn SH, Kim do Y, Shin JS, Kim YS, Hong SP, Chung HJ, Kim SO, Yoo WD, Han KH. Association between CCR5 promoter polymorphisms and hepatitis B virus infection. Korean J Hepatol. 2005. 11:116–124.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download