Abstract
The metastasis-suppressing role of the nm23 gene in the metastatic spread of malignant tumor is still debated. We examined the nm23-H1 protein expression and gene mutation in non-Hodgkin's lymphomas to compare with the clinicopathologic parameters. The expression of nm23-H1 protein was immunohistochemically examined in 150 cases of non-Hodgkin's lymphomas; 85 diffuse large B cell lymphomas (DLBCL), 18 marginal zone B cell lymphomas (MZL), 3 mantle cell lymphomas, 25 peripheral T cell lymphomas, not otherwise specified (TCLNOS), and 19 NK/T cell lymphomas (NK/T). Eighty-one cases (58 DLBCL, 6 MZL, 4 TCLNOS, and 13 NK/T) were studied for nm23-H1 gene mutation in exon 1 to 5. The high expression of nm23-H1 protein was associated with the high IPI score (p=0.019) and the low survival rate of the patients (p=0.0039). The gene mutation of nm23-H1 was detected in 10.3% of DLBCL and 30.7% of NK/T; but none in MZL and TCLNOS. The mutation was found in exon 1 in 5 cases, exon 2 in two cases, exon 4 in one case and both exon 1 and 2 in two cases. Our results suggest that the expression of nm23-H1 protein can be used as a poor prognostic marker in non-Hodgkin's lymphomas, and the mutational change of gene may operate in the lymphomagenesis.
The nm23 gene was first classified as a metastasis suppressor gene on the basis of its reduced expression in several metastatic melanoma cell lines relative to nonmetastatic counterparts (1). Subsequently, the low expression of nm23-H1 protein has been described as being linked to an increased metastatic potential in breast carcinoma, hepatoma, and gastric carcinoma, implying its role at inhibiting metastasis in a variety of human cancers (2-4). In other tumors, such as neuroblastoma, pancreatic carcinoma, and head and neck carcinomas, surprisingly, the opposite trend has been reported (5-7).
Malignant lymphomas constitute a heterogenous group of disorders, comprising different histogenesis and variable clinical outcomes. The histologic types and clinical staging are well known as prognostic indicators. Nevertheless they cannot accurately predict a clinical outcome in an individual case. Thus, a continuous necessity for reproducible quantitative methods has been required to confer additional prognostic information and to guide the clinician in the selection of the most appropriate therapeutic approach.
The previous studies of nm23-H1 expression in malignant lymphomas revealed that overall survival rates were significantly lower in the patients with nm23-H1-positive lymphomas than in those with nm23-H1-negative lymphomas (8-12). There has been only one study describing no mutational change of nm23-H1 gene in malignant lymphomas (13). The aims of this study are to determine the significance of nm23-H1 protein expression as a prognostic indicator and to detect the frequency of nm23-H1 gene mutation in non-Hodgkin's lymphomas.
One hundred fifty non-Hodgkin's lymphomas were selected from the pathology file in the Department of Pathology, Anam and Guro Hospitals of Korea University, Korea. The tissues were routinely processed with 10% buffered formalin fixation and paraffin embedding. The diagnosis and classification of lymphomas were dependent upon the routine histologic sections and immunohistochemical staining. They were supplemented by TCRγ rearrangement study and followed by the WHO classification (14). The studied cases were 106 B cell lymphomas (85 diffuse large B cell lymphomas [DLBCL], 18 marginal zone B cell lymphomas [MZL], and 3 mantle cell lymphomas [MCL]), and 44 T cell lymphomas (25 peripheral T cell lymphoma, not otherwise specified [TCLNOS], and 19 NK/T cell lymphomas [NK/T]). Forty-two cases were nodal and 108 cases were extranodal. The extranodal sites were 44 gastrointestinal tracts, 17 nasal cavities, 15 Waldeyer's rings, 12 ocular adnexae, 3 salivary glands, 3 thyroid glands, 3 testes, and 11 others. The median age of patients was 53.8 yr (15-88 yr). Ninty-six patients were male and fifty-four patients female. The patients were classified according to the Ann Arbor clinical staging system; 49 stage I, 58 stage II, 28 stage III, and 15 stage IV. The patients were classified according to the International Prognostic Index (IPI); 105 low risk (0, 1), 33 intermediate risk (2, 3), and 12 high risk (4, 5). Twenty-seven patients died from malignant lymphoma between 1 month to 58 months (mean; 13.7 months). One hundred twenty-three patients survived, and the mean follow-up duration was 26.9 months (2-120). This investigation was performed after obtaining an informed consent from all the patients involved in our study.
All slides were reviewed, and the appropriate area was marked and selected for tissue microarray. Agar blocks for tissue microarray were prepared as follows; Agar (4 g) was dissolved into 100 mL distilled water and boiled in microwave for 2 min. It was solidified in the cast and routinely processed as a tissue sample. The agar was embedded in paraffin. The selected area of each case was holed by the punch biopsy tool (2 mm in diameter) and transplanted into the agar block, holed by the same biopsy tool. After all of the cases were reembedded into the agar block, the block was incubated at 37℃ for 30 min to mould the tissue in paraffin.
The expression of nm23-H1 protein was immunohistochemically stained on 4 µm thick section from microarray blocks by the use of LSAB method. Monoclonal antibody to nm23-H1 (1:100, Novocastra Laboratories Ltd, U.K.) was used as a primary antibody. Immunohistochemical staining results for nm23-H1 protein were classified according to the following criteria: (-), when less than 5% of tumor cells were positive; (1+), when 5-50% of tumor cells were positive; (2+), when more than 50% of the tumor cells were positive (Fig. 1).
Total 81 cases (58 DLBCL, 6 MZL, 4 TCLNOS, 13 NK/T) were available for mutation study. DNA was extracted from the formalin-fixed, paraffin-embedded tissue sections by the use of the standard method with proteinase K digestion and phenol/chloroform purification. The primers for PCR were designed to amplify exons 1-5 as follows; Exon 1: (forward) 5'gtctgaaaaacgtagcgccgg3', (reverse) 5'cttaggtttgaactccggctg 3', Exon 2: (forward) 5'gcttgagacggatgacgctgta3', (reverse) 5'caggttaatcacagtgttctcc3', Exon 3: (forward) 5'atgtccttagatggtttgggggt3', (reverse) 5'tttggtctcattcatggctgtat3', Exon 4: (forward) 5'gccacattttctgctgtgatt3', (reverse) 5'cccaaatccttgtggcaact3', Exon 5; (forward) 5'gtctaatgtccatggagcttc3', (reverse) 5'cagatggtcggggatggtaac3'. The total 20 µL reaction volume contained 3 µL template, 1 µL out of each oligonucleotide primer, 2 µL dNTP, 11 µL ddH2O, 0.2 µL Taq polymerase, and 2 µL 10 × PCR buffer with Mg2+. Cycling conditions were as follows; an initial penetration at 95℃ for 5 min, 40 cycles each at 94℃ for 1 min, at 56℃ for 30 sec, at 72℃ for 30 sec, followed by one cycle at 72℃ for 10 min. PCR products were visualized by electrophoresis in 1.8% (w/v) agarose gel. Then the products were subjected to 8% non-denaturation polyacrylamide gel electrophoresis and stained with silver nitrate. DNA sequencing was performed in the cases showing abnormal bands on PCR-SSCP by the use of the Automatic Sequencer ABI 3730 (Applied Biosystem, Foster city, CA, U.S.A.).
Statistical significance was evaluated by using Kruskal-Wallis test for independent groups. The survival curves were calculated by using the Kaplan-Meier method, and then compared with other prognostic variables by using a log-rank test. All statistical analyses were two-sided with a significance level of p≤0.05, and all analyses were performed by using the SPSS statistical software (version 10.0, SPSS Inc., Chicago, IL, U.S.A.).
The immunostaining for nm23-H1 protein in a reactive lymph node showed a positive reaction in the cytoplasm of centroblasts in germinal centers and transformed lymphocytes in the interfollicular area. The staining in lymphoma cells was also primarily cytoplasmic. Of the 150 non-Hodgkin's lymphomas, 58 (38.6%) cases were (2+) and 46 (30.7%) cases (1+) for nm23-H1 protein (Table 1). According to the histologic types, 45.9% of DLBCL, 16.7% of MZL, 66.7% of MCL, 28.0% of TCLNOS, and 36.8% of NK/T lymphomas were (2+). In MZL, there was a lower expression tendency of nm23-H1 protein than in other types without statistical significance. The age, sex, and the nodal or extranodal sites were not correlated with nm23-H1 protein expression (Table 2). The nm23-H1 protein expression was significantly correlated with IPI score (p=0.019) but not with the Ann Arbor clinical stage (Table 3).
The survival rates in T cell lymphomas were worse than in B cell lymphoma (p=0.0415, Table 4). The higher Ann Arbor clinical stages (p=0.0000, Table 4), and IPI scores (p=0.0000, Table 4) were significantly associated with poor outcomes. The patients with (2+) expression of nm23-H1 protein showed the lower survival rate than those with negative or (1+) expression (p=0.0039, Table 4, Fig. 2). The high expression of nm23-H1 protein was associated with poor survival rates in B and T cell lymphomas (p=0.0013, Fig. 3). Even in the same Ann Arbor clinical stages (stage I, II and stage III, IV), and IPI groups (low, intermediate, and high), the patients with (2+) expression of nm23-H1 protein showed the lower survival rate than those with negative or (1+) expression (p=0.0042 and p=0.0483, Fig. 4).
By PCR-SSCP technique, the mutations were detected in 6 cases (10.3%) out of DLBCL and 4 cases (30.7%) out of NK/T. No mutation was found in MZL and TCLNOS (Table 5). These mutations were not correlated with age (p=0.071), sex (p=0.813), nodal or extranodal sites (p=0.738), Ann Arbor clinical stage (p=0.412), IPI (p=0.239), and survival rate (p= 0.1208). The mutation sites were as follows; five cases in exon 1, two cases in exon 2, two cases in exon 1 and 2, and one case in exon 4. Sequencing was performed in eight out of ten cases. All of the four cases with exon 1 mutation showed C→G transversion at nucleotide 55 (gi 468541) (Fig. 5). Three out of four cases with mutation in exon 2 showed insertional mutation after the nucleotide at 2257 (gi 468541) and the other one showed insertional mutation after the nucleotide at 2084 (Fig. 5). There was a weak correlation between the mutations and protein expression of nm23-H1 (p=0.057).
The human protein nm23-H1 is one of the human homologues of Drosophila awd (15). Eight different genes of this family have been identified in human, and their expressions are linked to suppression of tumor metastasis, differentiation, apoptosis, and proliferation (16,17). nm23-H1 and -H2 are nucleoside diphophate kinase, and metastasis suppression was observed in several tumor cell lines transfected with nm23-H1 gene (18,19). However, the biochemical mechanisms underlying the metastasis-suppressing action of nm23-H1 are largely unknown. The nm23 proteins are multifunctional, and they interact with many different proteins, some of which are involved in cancer progression (20). Indeed, the amount of nm23 increased in normal lymphocytes in response to mitotic stimulation and paralleled the increase in DNA synthesis (21). Our results also demonstrate the high expression of nm23-H1 protein is associated with poor prognosis. Therefore, the nm23 protein has a different and possibly varied role in tumors of different origins.
Niitsu et al. (11,12) reported that serum levels of nm23-H1 in aggressive non-Hodgkin's lymphomas were significantly higher than those in controls, and that high nm23-H1 levels were correlated with poor prognosis. Moreover, Niitsu et al. (8,9) reported 58.1% (100/172 patients) of DLBCL, 33.3% (5/15 patients) of MZL, 66.7% (12/18 patients) of MCL, 25.5% (12/47 patients) of TCLNOS, and 42.9% (9/21 patients) of NK/T were strongly positive (more than 70%) for nm23-H1. The survival rates were significantly lower in the patients with nm23-H1-positive lymphomas than in those with nm23-H1-negative lymphomas.
In our study, the degree of nm23-H1 protein expression was significantly correlated with IPI, but not with the age, sex, nodal or extranodal sites, and histologic types. The strong expression of nm23-H1 protein was significantly correlated with poor survival rate. Even in the same Ann Arbor clinical stage, IPI and histologic type (B and T cell lymphomas), the high expression of nm23-H1 protein was significantly correlated with poor survival rates. The results of this study were similar to the previous reports of Niistu et al. (8,9).
Mutational events of nm23 have not been frequently observed in human cancers, and its role in malignancy is controversial (22). Some data on colon and ovarian cancers suggested that mutations of nm23-H1 gene were associated with the presence of metastases, advanced disease, and poor survival (23,24). In neuroblastoma, a particular point mutation (Ser 120 Gly) affecting the protein folding was related to the aggressiveness (25). However, other groups were unable to detect any mutation of nm23-H1 gene in liver metastases of colon cancer, breast cancer, and hepatocellular carcinoma (3, 26-28). In malignant lymphomas, the mutational study by Aryee et al. (13) did not show any mutations by PCR-SSCP method.
In this study, we evaluated the nm23-H1 gene mutation by PCR-SSCP method and sequencing. We detected nm23-H1 mutation in non-Hodgkin's lymphoma. Ten out of eighty-one cases showed nm23-H1 mutations by PCR-SSCP method. These mutations were not correlated with clinicopathologic parameters. However, interestingly enough, the mutation was only found in DLBCL and NK/T lymphomas, but not in other types. In addition, the point mutation of exon 1 and insertional mutation of exon 2 were detected at the same sites; C to G transversion at nucleotide 55 of exon 1, and insertional mutation after the nucleotide at 2257. Bafico et al. (29) reported that a common polymorphic sequence (C to T transition, 30 nucleotides upstream from the 5'splice site of exon 1) in colorectal cancers, and Cipollini et al. (22) found that nm23-H1 gene mutation (C to A transversion at nucleotide -79 of exon 1) in breast cancers. The same sequence variants as the present study have not been described so far.
Because nm23-H1 protein can be easily examined at the time of conventional phenotypic examinations for the diagnosis of lymphoma, it may be used as a prognostic marker of non-Hodgkin's lymphoma in addition to known prognostic indicators. We found the new mutation sites in nm23-H1 gene. The role of these mutations in non-Hodgkin's lymphoma needs further investigation.
Figures and Tables
Fig. 1
The nm23-H1 protein expression by LSAB method. (A) (-), <5% positive cells; (B) (1+), 5-50% positive cells; (C) (2+), >50% positive cells.
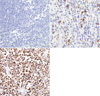
Fig. 2
The survival curve according to nm23-H1 protein expression (p=0.0039). -, less than 5% of tumor cells were positive; 1+, 5-50% of tumor cells were positive; 2+, more than 50% of the tumor cells were positive.

Fig. 3
The survival curve of nm23-H1 protein expression in B cell lymphoma (A) and T cell lymphoma (B) (p=0.0013). -, less than 5% of tumor cells were positive; 1+, 5-50% of tumor cells were positive; 2+, more than 50% of the tumor cells were positive.

Fig. 4
The survival curve of nm23-H1 protein expression according to the same Ann Arbor clinical stage (A, Stage III and IV, p=0.0042) and the same International Prognostic Index (B, IPI 2 and 3, p=0.0483).
-, less than 5% of tumor cells were positive; 1+, 5-50% of tumor cells were positive; 2+, more than 50% of the tumor cells were positive.

Fig. 5
nm23-H1 mutation by sequencing. (A) All of four cases with exon 1 mutation show the point mutation at the same site (*, C ← G). (B) Three of four cases with exon 2 mutation reveal the insertional mutation at the same site (*).

Table 1
The nm23-H1 protein expression according to the histologic types

DLBCL, Diffuse large B cell lymphoma; MZL, Marginal zone B cell lymphoma; MCL, Mantle cell lymphoma; TCLNOS, Peripheral T cell lymphoma, not otherwise specified; NK/T, NK/T cell lymphoma. -, less than 5% of tumor cells were positive; 1+, 5-50% of tumor cells were positive; 2+, more than 50% of the tumor cells were positive. p=0.229.
Table 3
The nm23-H1 protein expression according to the Ann Arbor stage (Stage) and the International Prognostic Index (IPI)
References
1. Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988. 80:200–204.

2. Mao H, Liu H, Fu X, Fang Z, Abrams J, Worsham MJ. Loss of nm23 expression predicts distal metastases and poorer survival for breast cancer. Int J Oncol. 2001. 18:587–591.

3. Fujimoto Y, Ohtake T, Nishimori H, Ikuta K, Ohhira M, Ono M, Kohgo Y. Reduced expression and rare genomic alteration of nm23-H1 in human hepatocellular carcinoma and hepatoma cell lines. J Gastroenterol. 1998. 33:368–375.

4. Kim YJ, Lee JH, Kim HR, Kim DY, Kim SK, Kim KK, Park S. Expression of the nm23 gene in primary and metastatic gastric cancer tissues. J Korean Surg Soc. 1999. 57:836–842.
5. Leone A, Seeger RC, Hong CM, Hu YY, Arboleda MJ, Brodeur GM, Stram D, Slamon DJ, Steeg PS. Evidence for nm23 RNA overexpression, DNA amplification and mutation in aggressive childhood neuroblastomas. Oncogene. 1993. 8:855–865.
6. Nakamori S, Ishikawa O, Ohigashi H, Imaoka S, Sasaki Y, Kameyama M, Kabuto T, Furukawa H, Iwanakga T, Kimura N. Clinicopathological features and prognostic significance of nucleoside diphosphate kinase/nm23 gene product in human pancreatic exocrine neoplasms. Int J Pancreatol. 1993. 14:125–133.
7. Pavelic K, Kapitanovic S, Radosevic S, Bura M, Seiwerth S, Pavelic LJ, Unusic J, Spaventi R. Increased activity of nm23-H1 gene in squamous cell carcinoma of the head and neck is associated with advanced disease and poor prognosis. Mol Med. 2000. 78:111–118.
8. Niitsu N, Nakamine H, Okamoto M, Akamatsu H, Higashihara M, Honma Y, Okabe-Kado J, Hirano M. Clinical significance of intracytoplasmic nm23-H1 expression in diffuse large B-cell lymphoma. Clin Cancer Res. 2004. 10:2482–2490.

9. Niitsu N, Nakamine H, Okamoto M, Akamatsu H, Honma Y, Higashihara M, Okabe-Kado J, Hirano M. Adult Lymphoma Treatment Study Group, ALTSG. Expression of nm23-H1 is associated with poor prognosis in peripheral T-cell lymphoma. Br J Haematol. 2003. 123:621–630.

10. Niitsu N, Honma Y, Iijima K, Takagi T, Higashihara M, Sawada U, Okabe-Kado J. Clinical significance of nm23-H1 proteins expressed on cell surface in non-Hodgkin's lymphoma. Leukemia. 2003. 17:196–202.

11. Niitsu N, Okabe-Kado J, Kasukabe T, Yamamoto-Yamaguchi Y, Umeda M, Homma Y. Prognostic implications of the differentiation inhibitory factor nm23-H1 protein in the plasma of aggressive non-Hodgkin's lymphoma. Blood. 1999. 94:3541–3550.
12. Niitsu N, Okabe-Kado J, Okamoto M, Takagi T, Yoshida T, Aoki S, Hirano M, Honma Y. Serum nm23-H1 protein as a prognostic factor in aggressive non-Hodgkin lymphoma. Blood. 2001. 97:1202–1210.

13. Aryee DN, Simonitsch I, Mosberger I, Kos K, Mann G, Schlogl E, Potschger U, Gadner H, Radaszkiewicz T, Kovar H. Variability of nm23-H1/NDPK-A expression in human lymphomas and its relation to tumour aggressiveness. Br J Cancer. 1996. 74:1693–1698.

14. Jaffe ES, Haris NL, Stein H, Vardiman JW, editors. Pathology and Genetics: Tumours of Haematopoietic and Lymphoid Tissues (World Health Organization Classification of Tumours). 2001. Lyon: IARC Press.
15. Biggs J, Hersperger E, Steeg PS, Liotta LA, Shearn A. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell. 1990. 63:933–940.

16. Lombardi D, Lacombe ML, Paggi MG. nm23: unrevealing its biological function in cell differentiation. J Cell Physiol. 2000. 182:144–149.
17. de la Rosa A, Williams RL, Steeg PS. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. Bioessays. 1995. 17:53–62.

18. Leone A, Flatow U, King CR, Sandeen MA, Margulies IM, Liotta LA, Steeg PS. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell. 1991. 65:25–35.
19. Russell RL, Pedersen AN, Kantor J, Geisinger K, Long R, Zbieranski N, Townsend A, Shelton B, Brunner N, Kute TE. Relationship of nm23 to proteolytic factors, proliferation and motility in breast cancer tissues and cell lines. Br J Cancer. 1998. 78:710–717.

20. Lombardi D, Mileo AM. Protein interactions provide new insight into Nm23/nucleoside diphosphate kinase functions. J Bioenerg Biomembr. 2003. 35:67–71.
21. Keim D, Hailat N, Melhem R, Zhu XX, Lascu I, Veron M, Strahler J, Hanash SM. Proliferation-related expression of p19/nm23 nucleoside diphosphate kinase. J Clin Invest. 1992. 89:919–924.

22. Cipollini G, Moretti A, Ghimenti C, Viacava P, Bevilacqua G, Caligo MA. Mutational analysis of the NM23.H1 gene in human breast cancer. Cancer Genet Cytogenet. 2000. 121:181–185.

23. Wang L, Patel U, Ghosh L, Chen HC, Banerjee S. Mutation in the nm23 gene is associated with metastasis in colorectal cancer. Cancer Res. 1993. 53:3652.
24. Mandai M, Konishi I, Komatsu T, Mori T, Arao S, Nomura H, Kanda Y, Hiai H, Fukumoto M. Mutation of the nm23 gene, loss of heterozygosity at the nm23 locus and K-ras mutation in ovarian carcinoma: correlation with tumour progression and nm23 gene expression. Br J Cancer. 1995. 72:691–695.

25. Lascu I, Schaertl S, Wang C, Sarger C, Giartosio A, Briand G, Lacombe ML, Konrad M. A point mutation of human nucleoside diphosphate kinase A found in aggressive neuroblastoma affects protein folding. J Biol Chem. 1997. 272:15599–15602.

26. Heide I, Thiede C, Poppe K, de Kant E, Huhn D, Rochlitz C. Expression and mutational analysis of Nm23-H1 in liver metastases of colorectal cancer. Br J Cancer. 1994. 70:1267–1271.

27. Callahan R, Cropp C, Sheng ZM, Merlo G, Steeg P, Liscia D, Lidereau R. Definition of regions of the human genome affected by loss of heterozygosity in primary human breast tumors. J Cell Biochem Suppl. 1993. 17G:167–172.

28. Cropp CS, Lidereau R, Leone A, Liscia D, Cappa AP, Campbell G, Barker E, Le Doussal V, Steeg PS, Callahan R. NME1 protein expression and loss of heterozygosity mutations in primary human breast tumors. J Natl Cancer Inst. 1994. 86:1167–1169.

29. Bafico A, Varesco L, De Benedetti L, Caligo MA, Gismondi V, Sciallero S, Aste H, Ferrara GB, Bevilacqua G. Genomic PCR-SSCP analysis of the metastasis associated NM23-H1 (NME1) gene: a study on colorectal cancer. Anticancer Res. 1993. 13:2149–2154.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download