Abstract
The M protein and streptococcus pyrogenic exotoxin (SPE A) are important virulence factors in group A streptococci (GAS) infections. The emm types of GAS strains isolated from patients with sepsis were determined by sequencing the 5' N-terminus of the emm gene, encoding the M protein, and clonality analysis using pulsed-field gel electrophoresis. The presence of speA and production of SPE A were also examined. There were no predominant GAS clones. The emm genotypes were variable, and the most common genotype was emm13 (17.9%). The production prevalence of SPE A was 21.4%. The low mortality rate (7.1%) of GAS sepsis might be attributable to the low incidence of virulent strains such as emm1 (10.7%) and emm3 (7.1%), as well as to low production rate of SPE A.
There has been an increase in the number of reported cases of group A streptococci (GAS) sepsis recently (1). The number of severe GAS infections, such as toxic shock-like syndrome (TSLS), also has increased in many countries, reflecting changing epidemiology and clinical patterns of invasive streptococcal infections (2-5). International communications about disease and strain typing are important in defining and monitoring trends in infectious diseases (6). The M protein and streptococcus pyrogenic exotoxin (SPE A) are the most important virulence factors in GAS infections. An increasing frequency of M serotypes 1 and 3 has been observed in the United States (7) and Europe (8). These types are more often involved in invasive and fatal infections than are other M serotypes (2, 9).
Although M typing has long been used for epidemiologic purpose, it is difficult to perform routinely because it is hard to produce and maintain more than 80 kinds of anti-M sera. Recently, genetic typing of GAS became available through the sequencing of the 5' N-terminal end of the emm gene, which encodes the M protein (10, 11). However, emm typing might not adequately reflect the genetic diversity of bacterial strains, as suggested by the finding that strains expressing the same M serotype can be distinguished by pulsed-field gel electrophoresis (PFGE) (6, 12).
In this work, genetic variability of GAS strains isolated from patients with sepsis was determined by emm genotyping and PFGE. Restriction-fragment length polymorphism (RFLP) analysis of emm PCR products was applied for the simple discrimination of each strain. The presence of speA and the production of SPE A were also examined to estimate the positive rate of this toxin in GAS sepsis.
A total of 28 GAS strains isolated from blood cultures from 1992 through 1999 were collected from four university hospitals: Seoul National University Hospital (17 strains), Asan Medical Center (4 strains), Ewha University Mokdong Hospital (1 strain), all located in Seoul, and Gyeongsang National University Hospital (6 strains), located in Jinju. The medical records were reviewed for all patients, with note taken of age, sex, underlying diseases, the presence of TSLS, and the clinical outcome. There were 28 patients with a median age of 51 yr (range 1 month-74 yr), of whom 38.5% were 15 yr or younger and 23.1% were older than 60 yr. Nine patients had cancer. The sources of sepsis were cellulitis in 4 cases, pneumonia in 3, and peritonitis in 2. In 11, no focus of infection could be identified. All but 2 patients of TSLS survived (Table 1).
For identification of GAS, β-hemolytic colonies on sheep blood agar plate were tested with bacitracin, latex agglutination (Strepto LA kit; Denka Seiken, Tokyo, Japan), and the VITEK GPI card (bioMeriux, Hazelwood, MO, U.S.A.).
All 28 isolates were tested for T type by slide agglutination (Sevac AS, Prague, Czech Republic).
The emm sequence analysis method was modified from that of Beall et al. (11). Forward and backward primers were used in PCR as described previously (12). The product was sequenced using primer 5'-TATTCGCTTAGAAAATTAAAAACAGG-3' (13) with dye-labeled terminator PCR mix and subjected to automated sequence analysis on a 373 Autosequencer (Applied Biosystems, Foster City, CA, U.S.A.). The DNA sequences were subjected to homology searches against the bacterial DNA database with the BLAST program from the U.S. National Center for Biotechnology Information. The corresponding emm sequence diverged by less than 4% over 150 to 200 bases.
The emm PCR products were cut with HaeIII and separated on 12% polyacrylamide gel for 2 hr at 15 mA. Gels were stained with ethidium bromide and photographed (Polaroid 667 film) under UV light.
Macrorestriction and PFGE were performed as previously described with several modifications (2). Genomic DNA was digested with SmaI or ApaI for 6 hr at 25℃. Fragments of DNA were separated on 1% pulsed field-certified agarose (Bio-Rad, Hercules, CA, U.S.A.) in 0.5× TBE buffer using a CHEF-Mapper XA system (Bio-Rad) for 27 hr with a 2.16- to 26.29-sec switch time and 74% ramping factor between 10 and 700 kb at a 120° angle and 6 V/cm. Gels were stained with ethidium bromide, rinsed in distilled water, and photographed under UV light.
The PCR primers were used for detection of speA as previously described (16). Initial denaturation of DNA at 94℃ for 3 min was followed by 30 cycles of 94℃ for 1 min, 50℃ for 1 min, and 72℃ for 1 min. The amplicon size for speA was 205 bp.
The bacterial suspension was precipitated with ethanol, mixed with loading buffer, and boiled for 5 min. The denatured protein was separated on 10% polyacrylamide gel for 1.5 hr at 16 mA. The gel was transferred to the nitrocellulose membrane for 15 hr at 80 mA, and the membrane was treated with anti-SPE A antibody. Color development was observed with anti-rabbit antibody and 4-chloro-1-naphthol.
There were three (10.7%) strains each of the T1, T12, and T28 types (Table 2). Ten strains (35.7%) were nontypeable. Two strains showed a rare combination of T and emm types, namely SJ8 (T6, emm13) and SJ21 (T4, emm2).
The most frequent emm type was 13 (17.9%). The next most common types were emm12 (14.3%), emm1 (10.7%), and emm28 (10.7%). There were 13 emm-types (Table 2).
One to three small fragments were separated on the gel after treatment with HaeIII (Fig. 1A). All strains except emm6 were digested with HaeIII and were classified into seven types (Fig. 1B). The emm49, emm53, and SP2346 types demonstrated similar RFLP patterns (H1) with emm1. The emm 2, 22, and 28 types also showed similar RFLP patterns (H2). Otherwise, each emm type had a unique RFLP pattern (Table 2).
All but 4 of the 28 strains were digested with SmaI, showing 10 to 15 large fragments (Fig. 2A). Two strains of emm75, one of emm6, and one of emm12 were not digested with SmaI, whereas they were cut with ApaI (Fig. 2B). The emm13, 12, and 1 types showed some variance of PFGE pattern. Otherwise, each emm type had a unique PFGE pattern. Thirteen PFGE patterns were found by SmaI (Fig. 3). All strains were cut with ApaI, and the genetic diversity revealed by ApaI agreed with the result of SmaI digestion. Exceptionally SJ6 and SJ8 (emm13) showed 3 different fragment bands with ApaI, while they were the same with SmaI enzyme.
The presence of speA and SPE A toxin was found only in the emm1, emm3, and SP2346 strains (Table 2).
GAS have long been recognized as serious human pathogens that cause life-threatening illnesses such as sepsis (1), rheumatic fever (RF), necrotizing fasciitis (16), and TSLS (3-5). Sepsis cases attributable to GAS had arisen sporadically in Korea (3, 12). In the U.S.A., 64% of GAS sepsis cases between 1989 and 1990 were caused by M1 serotype, compared with 18% of those before 1979 (17). More than 80% of the GAS isolated from patients with severe disease produce SPE, as opposed to less than 20% of isolates causing less serious disease (17). The M1 and M3 strains may be more likely to cause invasive infections (8). The SPE A protein, an inherent virulence factor, plays an important role in TSLS (5-7, 18). The emm1 and emm3 strains producing SPE A represented only 17.9% of the isolates in this study, which may explain the good outcomes of sepsis.
Both T and M typing have been used for surveillance and other epidemiologic studies. As anti-T sera have become commercially available, T typing has been widely used for screening for epidemiologic studies of GAS infections (14). We performed T typing, expecting high typeability, as we had already experienced more than 90% typeability for GAS strains isolated from throat cultures (19). However, the proportion of T-nontypeable strains was so high (35.7%) in this study that T typing was only partly helpful in classifying the isolates. Rare combinations of T and emm type were observed in two strains (T6, emm13; T4, emm2) (14, 20). The M1 and M3 strains are known to have caused the recent outbreaks of RF and TSLS (2, 4-7, 17). Although the study period is different, the previous studies of the prevalence of M types in school children showed that M1 (1.2%) and M3 (4.9%) were rare, while M12 (48.8%) and M5 (14.6%) were very common (n=82) in Seoul. The M1 (3.9%) and M3 (1.3%) strains were likewise rare, while M12 (26.3%), M22 (14.5%), and M28 (10.5%) were frequent (N=76) in Jinju (19). Although emm13 was the most common type in this study, it was not found in school children in Seoul and was recovered only from 2.6% of school children in Jinju (19). Another study revealed emm1 (20.1%) and emm3 (7.7%) were not uncommon in the clinical isolates of GAS in Seoul (12). No emm13 was identified among 194 clinical isolates (12). The authors demonstrated the difference of emm distribution between school children and the patients with acute pharyngitis in the same area (21). The M types corresponding to emm53 and SP2346 have not been identified previously in Korea (19, 22). An emm sequence analysis has the advantage of being able to differentiate serologically M-nontypeable strains and to identify new emm genes (11).
The emm-RFLP technique is easy and economical for epidemiologic studies (15) and is a rapid method of sorting large numbers of diverse GAS into distinct genotypes. The emm-RFLP method can be used as an alternative to emm sequence typing or PFGE in a small laboratory. The emm-RFLP analysis is applicable to areas where GAS are endemic and where the majority of isolates are M nontypeable by serotyping (23). Simple discrimination of the strains into seven groups was possible by emm-RFLP. Identical emm restriction cleavage profiles had the same emm gene sequences except for a few types. The emm49, emm53, SP2346 strain could not be differentiated from emm1 by emm-RFLP, as is also true of emm2, emm22, and emm28. Two strains of the emm6 type were not cut with HaeIII, which suggests the limitation of emm-RFLP for epidemiologic studies.
It is notable that the clones of the same emm types were maintained over different areas/hospitals for a long periods of time. Although there was no predominant clone, six small clusters of clones (emm 1, 3, 4, 12, 13, and 28) were found by PFGE (Table 2). Differences in PFGE patterns were noted within the same emm13 type. Two strains of emm75 and one each of emm6 and emm12 were not cut by SmaI despite repeated experiments. These strains were, however, digested with ApaI, suggesting DNA integrity during the PFGE procedure. Recently, there have been reports that strains carrying the macrolide resistance gene, mefA, are not cut with SmaI (24, 25). This gene may encode a SmaI site-modifying activity (25). Those strains uncut with SmaI in this study were confirmed to have the mefA gene by PCR (unpublished data).
The SmaI digests of chromosomal DNA of two emm3 strains appeared to have PFGE profiles identical to that of the PFGE-1 clone found in a study of invasive streptococcal disease in Minnesota (26). Although PFGE-1 clone serotyped M3 predominated in TSLS in the U.S.A. (26) and in Japan (27), it represented only 7.1% of our isolates. Notably one emm3 strain (SJ17) caused TSLS in this study. On the other hand, the findings of Chaussee et al. (28) did not support a clonal basis for the resurgence of invasive streptococcal infections by observing the genetic heterogeneity using PFGE. There might be no predominant emm type or PFGE pattern, because the GAS strains had been isolated sporadically over several years at four hospitals in this study. Drawbacks of this study are lack of evaluation of invasiveness or virulence of GAS isolates and limited number of cases. Following studies with enough number of cases are needed.
In conclusion, there were several small clusters of clones found among the GAS strains causing sepsis by T typing, emm, emm-RFLP and PFGE profiles. The emm types of the GAS isolated were variable, and the most common type was emm13. The production of SPE A was associated with emm1 and emm3 types. The proportion of SPE A-producing GAS was relatively low, which may explain the good prognosis of GAS sepsis in this study.
Figures and Tables
 | Fig. 1(A) emm-RFLP restricted with HaeIII enzyme runned on 12% acrylamide gel and (B) its schematic pattern. Number of each lane in (B) represents emm RFLP pattern. Lane M, molecular marker of 100 bp ladder. |
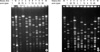 | Fig. 2Genomic DNA fingerprints by pulsed-field gel electrophoresis after digestion with (A) SmaI and (B) ApaI enzymes. Lane M, molecular marker of low range PFG. |
 | Fig. 3Schematic pattern of each representative fingerprints produced by SmaI enzyme. Marker, molecular marker of low range PFG. |
Table 1
Clinical features of 28 cases of group A streptococcal bacteremia

Hospitals: A, Seoul National University Hospital; B, Asan Medical Center; C, Ehwa Woman's University Mokdong Hospital; D, Gyeongsang National University Hospital.
LC, liver cirrhosis; PLCa, primary liver cancer; SLE, systemic lupus erythematosus; FUO, fever of unknown origin; SCLCa, small-cell lung cancer; AGCa, advanced gastric cancer; IUFD, intrauterine fetal death; DoD, died of streptococcal disease; TSLS, toxic shock-like syndrome; DM, diabetes mellitus; ALL, acute lymphocytic leukemia.
ACKNOWLEDGMENTS
The authors thank Hwan-Jong Lee (Seoul National University), Chik-Hyun Pai and Mi-Na Kim (University of Ulsan), and Mi-Ae Lee (Ewha Woman's University) for donating precious bacterial strains; Jun-Ho Oh for his excellent technical assistance; and Michio Ohta (Nagoya University, Nagoya, Japan) for giving us anti-SPE A sera.
References
1. Wheeler MC, Roe MH, Kaplan EL, Schlievert PM, Todd JK. Outbreak of group A streptococcus septicemia in children. JAMA. 1991. 266:533–537.

2. Kiska DL, Thiede B, Caracciolo J, Jordan M, Johnson D, Kaplan EL, Gruninger RP, Lohr JA, Gilligan PH, Denny FW Jr. Invasive group A streptococcal infection in North Carolina: epidemiology, clinical features, and genetic and serotype analysis of causative organisms. J Infect Dis. 1997. 176:992–1000.
3. Lee SY, Lee JS, Lee MA, Jung WS, Kim SJ. Streptococcal toxic shock syndrome associated with intrauterine fetal death: a case report. Korean J Clin Microbiol. 1998. 1:109–112.
4. Nakashima K, Ichiyama S, Iinuma Y, Hasegawa Y, Ohta M, Ooe K, Shimizu Y, Igarashi H, Murai T, Shimokata K. A clinical and bacteriologic investigation of invasive streptococcal infections in Japan on the basis of serotypes, toxin production, and genomic DNA fingerprints. Clin Infect Dis. 1997. 25:260–266.

5. Hoge CW, Schwartz B, Talkington D, Brieman RF, MacNeill EM, Englender SJ. The changing epidemiology of invasive group A streptococcal infections and the emergence of toxic shock-like syndrome. JAMA. 1993. 269:384–389.
6. Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, Martin DR. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995. 63:994–1003.

7. Musser JM, Hauser AR, Kim MH, Schlievert PM, Nelson K, Selander RK. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive disease: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991. 88:2668–2672.
8. Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980-1990 and their association with disease. J Med Microbiol. 1993. 39:165–178.

9. Hauser AR, Stevenes DL, Kaplan EL, Schlievert PM. Molecular analysis of pyrogenic exotoxins from Streptococcus pyogenes isolates associated with toxic shock-like syndrome. J Clin Microbiol. 1991. 29:1562–1567.

10. Caparon MG, Scott JR. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987. 84:8677–8681.

11. Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996. 34:953–958.

12. Lee YH, Hwang KJ, Lee KJ, Bae SM, Kim KS. Genotype analysis of pyrogenic exotoxin and emm genes of Streptococcus pyogenes clinical isolates. J Bacteriol Virol. 2003. 33:277–283.
13. Whatmore AM, Kehoe MA. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in Vir regulons. Mol Microbiol. 1994. 11:363–374.

14. Beall B, Facklam R, Hoenes T, Schwartz B. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J Clin Microbiol. 1997. 35:1231–1235.

15. Stanley J, Desai M, Xerry J, Tanna A, Efstratiou A, George R. High-resolution genotyping elucidates the epidemiology of group A streptococcus outbreaks. J Infect Dis. 1996. 174:500–506.

16. Kim SJ, Kim MG, Hwang YS, Yang JW, Koo KH, Jeong ST. Two cases of acute necrotizing fasciitis due to Streptococcus pyogenes. Korean J Infect Dis. 1996. 28:185–190.
17. Schwartz B, Facklam RR, Breiman RF. Changing epidemiology of group A streptococcal infections in the USA. Lancet. 1990. 336:1167–1171.
18. Talkington DF, Schwartz B, Black CM, Todd JK, Elliott J, Breiman RF, Facklam RR. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect Immun. 1993. 61:3369–3374.

19. Kim S. Bacteriologic characteristics and serotypings of Streptococcus pyogenes isolated from throats of school children. Yonsei Med J. 2000. 41:56–60.
20. Johnson DR, Kaplan EL. A review of the correlation of T-agglutination patterns and M-protein typing and opacity factor production in the identification of group A streptococci. J Med Microbiol. 1993. 38:311–315.

21. Kim S, Lee NY. Antibiotic resistance and genotypic characteristics of group A streptococci associated with acute pharyngitis in Korea. Microb Drug Resist. 2004. 10:300–305.

22. Kim S, Lee NY. Epidemiology and antibiotic resistance of group A streptococci isolated from healthy schoolchildren in Korea. J Antimicrob Chemother. 2004. 54:447–450.

23. Gardiner DL, Goodfellow AM, Martin DR, Sriprakash KS. Group A streptococcal Vir types are M-protein gene (emm) sequence type specific. J Clin Microbiol. 1998. 36:902–907.

24. Lee NY, Koh EH, Kim S. Clonality analysis using pulsed-field gel electrophoresis of erythromycin resistant group A streptococci. Korean J Clin Microbiol. 2004. 7:27–30.
25. Descheemaeker P, Chapelle S, Lammens C, Hauchecorne M, Wijdooghe M, Vandamme P, Ieven M, Goossens H. Macrolide resistance and erythromycin resistance determinants among Belgian Streptococcus pyogenes and Streptococcus pneumoniae isolates. J Antimicrob Chemother. 2000. 45:167–173.

26. Cockerill FR 3rd, Thompson RL, Musser JM, Schlievert PM, Talbot J, Holley KE, Harmsen WS, Ilstrup DM, Kohner PC, Kim MH, Frankfort B, Manahan JM, Steckelberg JM, Roberson F, Wilson WR. Molecular, serological and clinical features of 16 consecutive cases of invasive streptococcal disease. Southeastern Minnesota Streptococcal Working Group. Clin Infect Dis. 1998. 26:1448–1458.
27. Ichiyama S, Nakashima K, Shimokata K, Ohta M, Shimizu Y, Ooe K, Igarashi H, Murai T. Transmission of Streptococcus pyogenes causing toxic shock-like syndrome among family members and confirmation by DNA macrorestriction analysis. J Infect Dis. 1997. 175:723–726.

28. Chaussee MS, Liu J, Stevens DL, Ferretti JJ. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infection. J Infect Dis. 1996. 173:901–908.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download