Abstract
Complete or partial triplication of human chromosome 21 results in Down syndrome (DS). To analyze differential gene expressions in amniotic fluid (AF) cells of DS, we used a DNA microarray system to analyze 102 genes, which included 24 genes on chromosome 21, 28 genes related to the function of brain and muscle, 36 genes related to apoptosis, 4 genes related to extracellular matrix, 8 genes related to other molecular function and 2 house-keeping genes. AF cells were collected from 12 pregnancies at 16-18 weeks of gestation in DS (n=6) and normal (n=6) subjects. Our DNA microarray experiments showed that the expressions of 11 genes were altered by at least 2-folds in DS, as follows. Ten genes, COL6A1, CASP5, AKT2, JUN, PYGM, BNIP1, OSF-2, PRSS7, COL3A1, and MBLL were down-regulated and GSTT1 was only up-regulated. The differential expressions of GSTT1 and COL3A1 were further confirmed by semi-quantitative RT-PCR for each sample. The gene dosage hypothesis on chromosome 21 may explain the neurological and other symptoms of DS. However, our results showed that only two genes (COL6A1 and PRSS7), among 24 genes on chromosome 21, were down-regulated in the AF cells of DS. Our data may provide the basis for a more systematic identification of biological markers of fetal DS, thus leading to an improved understanding of pathogenesis for fetal DS.
Down syndrome (DS) has an incidence of one per 700 live births and is caused by complete or partial triplication of human chromosome 21 (1). Nondisjunction during meiosis may be responsible for the various phenotypes of DS, and -70% of nondisjunction errors have been found to occur during meiosis I and the other 30% during meiosis II (2, 3). DS is characterized clinically by abnormal facial and skeletal features (4), mental retardation (5), precocious dementia (6) and congenital heart disease (7, 8).
Recently, genomic approach for DS has been performed using various clinical samples. However, the gene expression study using DS fetus has some limitation, especially for amniotic fluid (AF) cells. AF is commonly used for prenatal diagnosis in obstetrics and gynecology, which includes various tissue of origin for fetal DS. Our genomic approach for AF cells of DS may provide basic information for understanding of the development of DS fetus.
The differential expression of genes located on the extra copy of chromosome 21 has been assumed to be responsible for the phenotypic abnormalities of DS, but this gene dosage hypothesis has not been fully assessed on a genome-wide basis. The expression patterns of genes related to phenotypic abnormalities of DS may provide insights into their potential roles in DS. For example, DSCR-1 gene on chromosome 21 is developmental regulator gene and is involved in neurogenesis. Moreover, its overexpression may contribute to brain abnormalities (9). SIM2 gene is a transcriptional regulator that operates as a important determinant of the central nervous system and is a candidate gene for the pathogenesis of mental retardation in DS (10). The overexpression of S100beta in the AF of DS fetuses may be related to the appearance of Alzheimer-type neuropathological changes in DS (11).
We used the DNA microarray technique (12) using AF cells in DS to investigate the pathogenesis of this syndrome, and also identify biological markers of DS in based on a high-throughput method. Although it is not fully clear how AF cell can represent the phenotype of this syndrome, the differential gene expression by extra copy of chromosome 21 would be expected even in AF cells, regardless of the tissue of origin (13). Our microarray analysis determined the expressions level of 102 genes potentially important in DS from cultured AF cells at 16-18 weeks of gestation.
AF samples of DS and normal subjects were collected from women undergoing routine amniocentesis for genetic testing. From July 2001 to November 2002, 12 women with pregnancies of DS (n=6) and normal (n=6) subjects, at the CHA General Hospital, College of Medicine, Pochon CHA University (Seoul, Korea), gave informed consent for the use of their AF cells, which only included for this study. Pochon CHA college of Medicine's Institutional Reviewed Board approved this study for human subjects. After centrifugation of AF (10 mL) at 1,800 rpm for 10 min, the pellet of AF 0.5 mL was added to 2 mL of CHANG media (IRVINE SCIENTIFIC, Santa Ana, CA, U.S.A.) in a culture dish. AF cells were grown in the tissue culture flask under 5% CO2 at 37℃ in CHANG media for DNA microarray analysis. Cytogenetic analysis for determination of DS was performed on metaphase spreads of cultured AF cells by standard method.
AF cells were obtained from patients at 16-18 weeks gestation. DS (n=6) and normal (n=6) subjects, respectively, total RNA was extracted from normal and DS AF cells (at 80-90% confluency) using RNeasy minikit (QIAGEN, Valencia, CA, U.S.A.). Total RNA isolated from every subject was quantitated. The purity of total RNA was confirmed by spectrophotometer and agarose gel electrophoresis. Each 30 µg of total RNA from DS (n=6) and normal (n=6) subjects was pooled, and labeled with either Cy3UTP or Cy5UTP (NEN Life Science Products, Boston, MA, U.S.A.) during reverse transcription (RT). The RT was performed using 2 µg/µL oligo dT (Invitrogen, Carlsbad, CA, U.S.A), 0.1 M DTT, 200 µ/µL superscript enzyme, 5× first strand buffer (Gibco BRL, Cergy Pontoise, France), 25 mM dATP, dGTP, dCTP, 15 mM dTTP (Amersharm, Pharmacia, Piscataway, NJ, U.S.A.), 1 mM Cy3 or Cy5 labeled dUTP (NEN). Reaction mixture was incubated at 65℃ for 10 min for denaturation, 42℃ for 2 hr for RT. After first strand cDNA synthesis the RNA was degraded by adding 15 µL of 0.1 N NaOH and incubating at 65℃ for 30 min. 15 µL of 0.1 N HCl was added for neutralization.
We used cDNA chip which contained 102 genes located on chromosome 21 (24 genes), genes expressed in brain (11 genes) or muscle (17 genes) and apoptosis related genes (36 genes), extracellular matrix (ECM) related genes (4 genes), genes related to other molecular function (8 genes) and housekeeping genes (2 genes). The list of genes with accession number was shown in Table 1, and functional category of total genes analyzed by GeneSpring was shown Fig. 1A (Silicon Genetics, Redwood City, CA, U.S.A.). PCR-ampilfied EST of 102 genes were fabricated by duplicate on Corning glass slide (Disgene, Seoul, Korea).
Fluorescent cDNA probes were dried after ethanol preparation, and resuspended in 20 µL hybridization buffer of TE (pH 8.0), 20× SSC and 10% SDS. The labeled cDNA was heated at 100℃ for 2 min then incubated at 37℃ for 30 min. Reaction mixture was dropped on the slide and covered by a cover slip. The slide was assembled with a hybridization chamber and hybridized for 16 hr at 65℃.
The slide was dried by centrifugation and then scanned on ScanArray 4000XL (Packard Bioscience, Billerica, MA, U.S.A.). After registration of Cy3 and Cy5 images, the unified image was quantified using ImaGene™ ver 4.0 (BioDiscovery, Inc., Los Angeles, CA, U.S.A.). Normalization was performed from the quantified data by mean intensity of 102 genes on the slide (Global normalization).
For confirmation of gene expression level, semi-quantitative RT-PCR was performed using OneStep RT-PCR kit (QIAGEN, Inc., Valencia, CA, U.S.A.) according to the manufacturer's direction. For RT-PCR, 100 ng of total RNA was reverse transcribed at 50℃ for 30 min in 50 µL final volume by 2 µL of OneStep RT-PCR enzyme mix. Each gene specific primers were added in first strand synthesis step. The RT-PCR products were subjected to electrophoresis on 1% agarose gel. Differentially expressed genes were detected using the following primer pairs; COL3A1 sense, 5'-gtggacagattctagtgctgag-3', antisense, 5'-ataggtagtctcacagccttgc-3', GSTT1 sense, 5'-tgactactggtaccctcaggac-3', antisense, 5'-aggtcagctaaggagatgtgag-3', and GAPDH sense, 5'-accacagtccatgccatcac-3', antisense, 5'-tccaccaccctgttgctgta-3'
Microarray is powerful tool for analyzing expression profile and identifying bio-marker in clinical samples. In our microarray experiment, the RNA samples of the normal (n=6) and DS (n=6) subjects were pooled to reduce the effect of individual variations. Hybridization spots were quantified and normalized using ImaGene™ ver 4.0 software (BioDiscovery, Inc., Marina del Rey, CA, U.S.A.). To normalize the intensity levels of genes on the chip, we compared the Cy3:Cy5 intensity ratios of all spots on the array (Global normalization). GAPDH and β-actin, the housekeeping genes, were used as controls and the signal intensity of these genes was represented the almost same in Cy3 and Cy5 (Fig. 1B). The overall hybridization signals obtained by using cDNA probe of normal and DS subjects were similar (<2-folds). Eleven genes in DS showed the differences of 2-folds or more (Fig. 2).
Differentially expressed genes are represented by a pair mean ratio (Log2Test Mean/Control Mean) of signal intensity. The overexpression of gene was defined as a pair mean ratio of >1.0. Eleven genes were differentially expressed in DS, as summarized in Table 2. Only one gene, GSTT1, was up-regulated (1/102, 1%) and 10 genes were down-regulated in DS (10/102, 10%); OSF-2, PYGM, AKT2, JUN, BNIP1, COL-3A1, COL6A1, CSAP5, PRSS7 and MBLL (Fig. 2). These genes fell into the following groups: 1) Cell communication; OSF-2 and PYGM, 2) Signal transduction; AKT2, JUN and BNIP1, 3) Apoptosis; CSAP5, 4) Enzyme; PRSS7 and GSTT1. The functional categorization differentially expressed genes were classified using GeneSpring (ver 6 Silicon Genetics, Redwood, CA, U.S.A.) and summarized in Table 2.
We confirmed the microarray results to validate these differentially expressed genes in DS and normal samples by semiquantitative RT-PCR, and the matched results were observed (data not shown). We selected two genes; one up-regulated gene (GSTT1) and one down-regulated gene (COL3A1), for analysis of every subject, and the semi-quantitative RT-PCR results are shown in Fig. 3. GSTT1 was highly expressed in 50% of DS subjects (3/6), but in only one of normal subjects (1/6, 17%). The expression of COL3A1 was down-regulated in the DS subjects (5/6, 83%), but not in the normal subjects (0/6, 0%).
The gene dosage hypothesis by extra chromosome 21 may explain the neurological and other symptoms of DS. We analyzed the differential gene expression of AF cells of DS, although it is not fully clear how our analysis using AF cells can lead to the syndrome phenotype. Our analysis using AF cells may not explain the direct pathogenesis of common DS phenotype, but could be important for prenatal diagnosis and the study of DS fetus development.
According to the "gene dosage effect" hypothesis, the differential regulation of chromosome 21 genes causes the DS phenotype. Gazzolo et al. reported that S100B protein level of AF were significantly higher (1.5 fold) in DS fetuses (14). S100B gene is located on chromosome 21 and is calcium binding protein originally isolated from the nervous system (15). This gene is not only overexpressed in AF, but also life-long overexpressed in DS (11).
In the present study, we identified 11 differentially expressed genes in AF cells in DS. GSTT1, the up-regulated gene, plays a role for the intracellular binding, transport of many bio-molecular entities, and detoxification process. GSTT1 was not detected in any fetal organs examined, but found in deciduas (16). Moreover, this gene was absent in 38% of the population (17), and we found this gene up-regulated in AF cells of DS by approximately 4-folds in semi-quantitative RT-PCR analysis.
COL6A1, COL3A1 and OSF-2, three of the ten down-regulated genes, code for components of the extracellular matrix (ECM). COL6A1 protein was reported to be down-regulated in the brain of DS fetus (18) and to be expressed in the developing atrioventricular (AV) canal. Genetic variations of this gene have been associated with DS AV defects in human genetic studies (19). According to recently published data, expression changes of the ECM-related genes may also contribute to cardiac defects (19) and abnormalities of brain in DS (20, 21). COL3A1 gene, which encodes chains of type III procollagen, is important for the development of skin, the cardiovascular system and maintaining the normal physiological functions of these organs (22). Superti-Furga et al. provided the first description of a mutation of the COL3A1 gene in type IV Ehlers-Danlos syndrome (EDS). Synthesis of type III collagen is defective in type IV EDS (23). OSF-2, transcription activation protein, might play a role in cell to cell communication in bone, ECM turnover and switches cells into the osteoblastic pathway (24). It is possible that abnormal expression of OSF-2 may affect increased flexibility in joints of DS patients.
Beta amyloid precursor protein (APP), CuZn superoxide dismutase (SOD1) and S100beta have been implicated in causing apoptosis thought to be responsible for neuronal loss in DS (25). Little is known, however, about the changes of caspases and their regulatory proteins in DS. Gulesserian et al. reported that procaspase-3 and -8 were significantly decreased in frontal cortex (26). Although we used AF cells in this study, caspase-3 was down-regulated. BNIP1, Pro-apoptotic protein, was also down-regulated. This gene interacts with BCL-2 family which is anti-apoptosis proteins (27).
According to the gene dosage hypothesis, some genes on chromosome 21 would overexpress in DS. Previous study showed that the genes located on chromosome 21 have been found to be overexpressed in cells and tissues of DS (28). Our data showed that the expression level of most genes located on chromosome 21 (22/24 genes) did not change in the AF cells of DS. Only two genes (COL6A1 and PRSS7) were differentially expressed, especially for down regulation in the AF cells of DS. Our microarray analysis determined the expressions of 102 genes potentially important in DS and may provide the basis for a more systematic identification of biomarkers, thus leading to understanding of the developmental and pathogenic study for fetal DS.
Figures and Tables
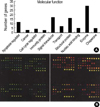 | Fig. 1The ontology of genes on the chip. (A) One hundred two genes were classified into functional subgroups using Simplified ontology in GeneSpring (Silicon Genetics, Redwood City, CA, U.S.A.). (B) The PCR amplicons of human cDNA clones were spotted in 4 blocks using a 4-pin print head, and all of genes spotted in duplicate. The similar result has shown by the repeated experiments. GAPDH and β-actin were spotted in last row of each blocks. |
 | Fig. 2Histogram and scatter plot by DNA microarray experiment. (A) This histogram shows the distribution of gene expression ratios between normal and DS subjects. One gene (GSTT1) was up-regulated and ten genes were down-regulated in DS. The horizontal axis refers to the expression ratio, and vertical axis indicates number of genes. (B) Each spot represents a single gene. Genes were expressed the same level in two samples fall along 45° angle line. Black spots were expressed over 2-folds but empty spots are expressed less than 2-folds. |
ACKNOWLEDGEMENT
This study was supported by a grant of the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (01-PJ10-PG6-01GN13-0002). The human cDNA clones on the chip were kindly provided from the Center for Functional analysis of Human Genome, KRIBB, Korea.
References
1. Korenberg JR, Chen XN, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, Graham JM Jr, Hudgins L, Mcgillivray B, Miyazaki K, Ogasawara N, Park JP, Pagon R, Pueschel S, Sack G, Say B, Schuffenhauer S, Soukup S, Yamanaka T. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994. 91:4997–5001.

2. Lamb NE, Feingold E, Savage A, Avramopoulos D, Freeman S, Gu Y, Hallberg A, Hersey J, Karadima G, Pettay D, Saker D, Shen J, Taft L, Mikkelsen M, Petersen MB, Hassold T, Sherman SL. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum Mol Genet. 1997. 6:1391–1399.

3. Antonarakis SE. 10 years of Genomics, chromosome 21, and Down syndrome. Genomics. 1998. 51:1–16.
4. Woodhouse JM, Hodge SJ, Earlam RA. Facial characteristics in children with Down's syndrome and spectacle fitting. Ophthalmic Physiol Opt. 1994. 14:25–31.

5. Engidawork E, Gulesserian T, Fountoulakis M, Lubec G. Aberrant protein expression in cerebral cortex of fetus with Down syndrome. Neuroscience. 2003. 122:145–154.

6. Takashima S, Becker LE, Armstrong DL, Chan F. Abnormal neuronal development in the visual cortex of the human fetus and infant with Down's syndrome. A quantitative and qualitative Golgi study. Brain Res. 1981. 225:1–21.

7. Ferencz C, Neill CA, Boughman JA, Rubin JD, Brenner JI, Perry LW. Congenital cardiovascular malformations associated with chromosome abnormalities: an epidemiologic study. J Pediatr. 1989. 114:79–86.

8. Freeman SB, Taft LF, Dooley KJ, Allran K, Sherman SL, Hassold TJ, Khoury MJ, Saker DM. Population-based study of congenital heart defects in Down syndrome. Am J Med Genet. 1998. 80:213–217.

9. Fuentes JJ, Genesca L, Kingsbury TJ, Cunningham KW, Perez-Riba M, Estivill X, de la Luna S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum Mol Genet. 2000. 9:1681–1690.

10. Yamaki A, Tochigi J, Kudoh J, Minoshima S, Shimizu N, Shimizu Y. Molecular mechanisms of human single-minded 2 (SIM2) gene expression: identification of a promoter site in the SIM2 genomic sequence. Gene. 2001. 270:265–275.

11. Griffin WS, Sheng JG, McKenzie JE, Royston MC, Gentleman SM, Brumback RA, Cork LC, Del Bigio MR, Roberts GW, Mrak RE. Life-long overexpression of S100beta in Down's syndrome: implications for Alzheimer pathogenesis. Neurobiol Aging. 1998. 19:401–405.
12. Roh MS, Hong SH, Jeong JS, Kwon HC, Kim MC, Cho SH, Yoon JH, Hwang TH. Gene expression profiling of breast cancers with emphasis of beta-catenin regulation. J Korean Med Sci. 2004. 19:275–282.
13. Prusa AR, Marton E, Rosner M, Freilinger A, Bernaschek G, Hengstschlager M. Stem cell marker expression in human trisomy 21 amniotic fluid cells and trophoblasts. J Neural Transm Suppl. 2003. 67:235–242.

14. Gazzolo D, Bruschettini M, Corvino V, Lituania M, Sarli R, Bruschettini P, Michetti F. Amniotic fluid levels of S100B protein in normal and trisomy-21 foetuses. Clin Chim Acta. 2003. 330:131–133.

15. Heizmann CW. Ca2+-binding S100 proteins in the central nervous system. Neurochem Res. 1999. 24:1097–1100.
16. Raijmakers MT, Steegers EA, Peters WH. Glutathione S-transferases and thiol concentrations in embryonic and early fetal tissues. Hum Reprod. 2001. 16:2445–2450.

17. Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994. 300(Pt 1):271–276.

18. Engidawork E, Baiic N, Fountoulakis M, Dierssen M, Greber-Platzer S, Lubec G. Beta-amyloid precursor protein, ETS-2 and collagen alpha 1 (VI) chain precursor, encoded on chromosome 21, are not overexpressed in fetal Down syndrome: further evidence against gene dosage effect. J Neural Transm Suppl. 2001. 61:335–346.
19. Davies GE, Howard CM, Farrer MJ, Coleman MM, Bennett LB, Cullen LM, Wyse RK, Burn J, Williamson R, Kessling AM. Genetic variation in the COL6A1 region is associated with congenital heart defects in trisomy 21 (Down's syndrome). Ann Hum Genet. 1995. 59(Pt 3):253–269.

20. Loftis MJ, Sexton D, Carver W. Effects of collagen density on cardiac fibroblast behavior and gene expression. J Cell Physiol. 2003. 196:504–511.

21. Anlar B, Atilla P, Cakar AN, Kose MF, Beksac MS, Dagdeviren A, Akcoren Z. Expression of adhesion and extracellular matrix molecules in the developing human brain. J Child Neurol. 2002. 17:707–713.

22. Olsen BR. The roles of collagen genes in skeletal development and morphogenesis. Experientia. 1995. 51:194–195.

23. Superti-Furga A, Gugler E, Gitzelmann R, Steinmann B. Ehlers-Danlos syndrome type IV: a multi-exon deletion in one of the two COL3A1 alleles affecting structure, stability, and processing of type III procollagen. J Biol Chem. 1988. 263:6226–6232.

24. Sugiura T, Takamatsu H, Kudo A, Amann E. Expression and characterization of murine osteoblast-specific factor 2 (OSF-2) in a baculovirus expression system. Protein Expr Purif. 1995. 6:305–311.

25. Engidawork E, Lubec G. Protein expression in Down syndrome brain. Amino Acids. 2001. 21:331–361.

26. Gulesserian T, Engidawork E, Yoo BC, Cairns N, Lubec G. Alteration of caspases and other apoptosis regulatory proteins in Down syndrome. J Neural Transm Suppl. 2001. 61:163–179.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download