Abstract
We report a Korean patient with glycogen storage disease type 1b (GSD-1b) whose diagnosis was confirmed by liver biopsy and laboratory results. The patient presented with delay of puberty and short stature on admission and had typical clinical symptoms of GSD as well as chronic neutropenia and inflammatory bowel disease. Mutation analysis of the glucose 6-phosphate translocase 6-phosphate translocase (SLC37A4) gene revealed that the patient was a compound heterozygote of two different mutations including a deletion mutation (c.1042_1043delCT; L348fs) and a missense mutation (A148V). The L348fs mutation was inherited from the patient's father and has been reported in an Italian family with GSD-1b, while the A148V mutation was transmitted from the patient's mother and was a novel mutation. To the best of our knowledge, this is the first report of genetically confirmed case of GSD-1b in Korean.
Glycogen storage disease type 1b (GSD-1b, MIM 232220) is an autosomal recessive disease caused by a deficiency of microsomal glucose-6-phosphate translocase (G6PT). This protein transports glucose-6-phosphate into the endoplasmic reticulum, where the enzyme glucose-6-phosphatase (G6Pase; EC 3.1.3.9) converts glucose-6-phosphate into glucose and inorganic phosphate. In 1997, Gerin et al. found that a gene coding for the G6PT was mutated in GSD-1b (1). This gene, SLC37A4 (formerly called as G6PT1), is located on chromosome 11q23.3 and encodes a protein with 429 amino acids (1, 2). Apart from GSD-1b, mutations in the SLC37A4 gene were also found in essentially all patients previously classified as GSD 1c (2, 3) and 1d (4). Owing to the discovery, GSD-1b, -1c, or -1d can be diagnosed by the direct mutation analysis of the SLC37A4 gene, which can spare the patients from invasive liver biopsy. In addition, the genetic causes in a family can facilitate family screening and is essential for prenatal diagnosis (5). Recently, we experienced a Korean patient with typical clinical features of GSD-1b and performed a mutation analysis to detect the SLC37A4 gene mutations. In this report, we present the result of the mutation analysis in this patient.
The proband was the first child of non-consanguineous Korean parents. He presented at age 12 yr for failure to thrive and protuberant abdomen. His height and weight was below the third percentile. Physical examination showed an enlarged liver, which was 7 cm below the costal margin. Fasting blood glucose was 40 mg/dL (reference interval: 70-110 mg/dL), blood lactate 5.6 mg/dL (reference interval: 0.7-2.5 mg/dL), serum uric acid 13.1 mg/dL (reference interval: 3.0-8.3 mg/dL), and serum triglyceride 586 mg/dL (reference interval: 50-200 mg/dL). Liver function was within normal limit except a mild elevation of serum ALT 65 U/L (reference interval: <40 U/L). A glucose-loading test (1.75 g/kg) resulted in rapid decrease in blood lactate and pyruvate. The neutrophil count of the peripheral blood ranged from 0.4×109/L to 0.8×109/L (reference interval: 1.5-9.0×109/L). He had experienced protracted diarrhea and recurrent perianal abscess. On colonoscopic finding, he had an inflammatory bowel disease. He underwent a liver biopsy in suspect of GSD. The biopsy showed an increased glycogen contents (12.3%; reference interval: 1-6%/wet liver), which is compatible to GSD. The G6Pase activities were measured with glucose-6-phosphate as substrate and were incubated in a reaction mixture containing 0.25 M saccharose and 5 mM EDTA buffer, pH 7.2. Microsome permeabilized with sonification and at least two G6P uptake studies were performed for each microsomal preparation. The patient's G6Pase activities in liver homogenates showed a very low level, 0.18 and 0.41 (normal range; 5.9-93.0 nM/min/mg protein) in intact microsome and disrupted microsome by sonification, respectively. Based on the clinical and laboratory findings, GSD-1b was considered. But G6Pase activity in disrupted microsome was a very low level, 0.41, but approximately approximately 2.3 times elevated level than that in untreated microsomes. In the first place, we had mutational analysis of the G6Pase gene to exclude GSD-1a and observed no mutation. We then analyzed mutations of SLC37A4 gene because we suspected clinically GSD-1b.
Genomic DNA was extracted from whole blood using a Wizard genomic DNA purification kit according to the manufacturer's instruction (Promega, Madison, WI, U.S.A.). Informed consent was obtained from the parents. All the exons and intron-exon boundaries of the SLC37A4 gene were amplified as described previously (3). Direct sequencing of PCR products was performed on both forward and reverse strands using the same primers for PCR and cycle sequencing was performed with a BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA, U.S.A.) on the ABI 3,100 Genetic Analyzer (Applied Biosystems). The sequence was compared with the reference sequence NM_001467 in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov).
Two different mutations in the SLC37A4 gene were identified in the proband. One was a heterozygous deletion mutation (c.1042_1043delCT; L348fsX400) in exon 8 and the other was a heterozygous 1-bp substitution mutation (c.443C>T; A148V) in exon 3 of the SLC37A4 gene. No other polymorphism was found in the sequencing results. The first mutation, c.1042_1043delCT, has been previously reported in an Italian family with GSD-1b (6). The second one is a novel mutation, c443C>T, leading to substitution of alanine by valine at codon 148 (A148V) (Fig. 1). The mother is heterozygous for the A148V mutation and the father is heterozygous for the L348fs mutation. The A148V mutation has been screened in 100 normal chromosomes and none had the same mutation.
GSD1 or von Gierke disease includes a clinically, biochemically, and genetically heterogeneous group of autosomal recessive disorders (7). The basic defects reside in the impairment of the terminal steps of glycogenolysis and gluconeogenesis, at different levels. Mutations of the glucose-6-phosphatase gene (G6Pase), which lead to the enzyme deficiency, are responsible for the most frequent form of GSD 1, the subtype 1a (7). This gene is not mutated in patients in which there are biochemical evidences of defects in the glucose-6-phosphate transport system (8). GSD 1 patients diagnosed as non 1a have been subdivided at least in 1b and 1c subtypes on the basis of clinical and biochemical parameters (7). In GSD -1a, the WBC count is generally within reference ranges because leukocyte function is unaffected by the defect. In contrast, GSD-1b causes chronic neutropenia due to the impaired function of the neutrophils, particularly in relation to Gram-positive organisms (9). In GSD-1b, a liver biopsy in which the hepatocytes and their microsomes are intact shows deficient G6Pase activity in enzyme assay, as the defects in the glucose-6-phosphate transport system dose not deliver the required substrate into the microsomal lumen. However, microsomes disrupted by solubilization show normal glucose-6-phosphatase activity, because all substrates have free entry to the enzyme. But G6Pase activity in disrupted microsome by sonification showed a very low level in our enzyme assay, which was not compatible finding to GSD-1b. Since liver biopsy specimen was not left, we could not try to repeat the G6Pase enzyme assay. Our G6Pase enzyme assay result may be caused by insufficient or inadequate sonification procedure for microsome disruption.
The recent cloning of the cDNA for a putative endoplasmic reticulum glucose-6-phosphate transporter (G6PT) (1) has enabled the search for mutations in this gene in non GSD-1a patients (4, 6, 10-13). Recent data from others (4, 10-13) indicate that mutations in the SLC37A4 gene are present in both 1b and 1c patients so far investigated. The SLC37A4 gene product is estimated to function as a transporter for a monophosphate ester by structural comparison to various types of bacterial monophosphate ester transporter proteins (1). The bacterial transporter proteins as well as a human G6P translocase have 12 transmembrane helices. The genomic structure showed that helix 1 is in exon 1, helices 2-4 in exon 2, helices 5 and 6 in exon 3, helix 7 in exon 4, helix 8 in exon 5, helix 9 in exon 6, helix 10 and the upper region of helix 11 in exon 7, and the lower region of helix 11 and helix 12 in exon 8, indicating close correlation between transmembrane helices and exons (14).
Based on genome sequence data, we analyzed DNA of a patient with GSD-1b. We identified two different mutations in Korean patient. The first mutation at the codon 347 cause a change in reading frame after Ala-347 and premature termination of translation in amino acid 400, which has been previously reported in Italian family with GSD-1b (6). This can possibly cause an abnormal folding of the transporter in the ER membrane and, consequently, a functional anomaly. On the other hand, nonsense mutations and insertion/deletion mutations cause the synthesis of a truncated protein mussing the two lysines at the carboxy-terminus necessary as a retention signal in the ER membrane and therefore do not allow for glucose-6-phosphate transport.
The second mutation at the codon 148, novel mutation was a missence mutation from alanine to valine at codon 148. This alanine locates in the third transmembrane helix. This amino acid change might alter the protein structure at the transmembrane region. All the known missence mutations causing amino acid substitutions in the transmembrane region of the protein are responsible for the conversion of a hydrophobic amino acid to a hydrophilic amino acid. Fifty normal subjects were screened by direct sequencing and none of them carried for the A148V mutation in the SLC37A4 gene. To determine whether these mutations were common to Korean GSD-1b patients, mutation screening of more patients should be undertaken.
In summary, we identified a novel missence mutations in the SLC37A4 gene in a Korean boy with the typical phenotype of GSD-1b, which we believe would improve our understanding of the genotype-phenotype correlations of SLC37A4 gene mutations. The DNA-based diagnosis of GSD-1b will enable us to make an accurate determination of carrier status and to perform prenatal diagnosis of this disease. To our knowledge, this is the first report of genetically confirmed case of GSD-1b in Koreans.
Figures and Tables
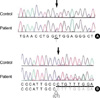 | Fig. 1Identification of SLC37A4 gene mutations. (A) Direct sequencing analysis demonstrated a heterozygous C to T transition (arrow; c.443C>T) resulting in a A148V missense mutation was observed in exon 3. (B) A heterozygous 2-bp deletion (arrow; c.1042-1043delCT) resulting in a A347fs×400 mutation in exon 8. Because the sequencing was performed with an anti-sense primer, overlapped peaks appear from the C+G peaks (arrow). |
References
1. Gerin I, Veiga-da-Cunha M, Achouri Y, Collet JF, Van Schaftingen E. Sequence of a putative glucose 6-phosphate translocase, mutated in glycogen storage disease type Ib. FEBS Lett. 1997. 419:235–238.
2. Veiga-da-Cunha M, Gerin I, Chen YT, de Barsy T, de Lonlay P, Dionisi-Vici C, Fenske CD, Lee PJ, Leonard JV, Maire I, McConkie-Rosell A, Schweitzer S, Vikkula M, Van Schaftingen E. A gene on chromosome 11q23 coding for a putative glucose-6-phosphate translocase is mutated in glycogen-storage disease types Ib and Ic. Am J Hum Genet. 1998. 63:976–983.
3. Galli L, Orrico A, Marcolongo P, Fulceri R, Burchell A, Melis D, Parini R, Gatti R, Lam C, Benedetti A, Sorrentino V. Mutations in the glucose-6-phosphate transporter (G6PT) gene in patients with glycogen storage diseases type 1b and 1c. FEBS Lett. 1999. 459:255–258.

4. Veiga-da-Cunha M, Gerin I, Chen YT, Lee PJ, Leonard JV, Maire I, Wendel U, Vikkula M, Van Schaftingen E. The putative glucose 6-phosphate translocase gene is mutated in essentially all cases of glycogen storage disease type I non-a. Eur J Hum Genet. 1999. 7:717–723.

5. Lam CW, Sin SY, Lau ET, Lam YY, Poon P, Tong SF. Prenatal diagnosis of glycogen storage disease type 1b using denaturing high performance liquid chromatography. Prenat Diagn. 2000. 20:765–768.

6. Marcolongo P, Barone V, Priori G, Pirola B, Giglio S, Biasucci G, Zammarchi E, Parenti G, Burchell A, Benedetti A, Sorrentino V. Structure and mutation analysis of the glycogen storage disease type 1b gene. FEBS Lett. 1998. 436:247–250.

7. Chen YT, Bruchell A. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Glycogen storage disease. The Metabolic and Molecular Bases of Inherited Disease. 7th edn. McGraw-Hill;395–965.
8. Lei KJ, Pan CJ, Shelly LL, Liu JL, Chou JY. Identification of mutations in the gene for glucose-6-phosphatase, the enzyme deficient in glycogen storage disease type 1a. J Clin Invest. 1994. 93:1994–1999.

9. Beaudet AL, Anderson DC, Michels VV, Arion WJ, Lange AJ. Neutropenia and impaired neutrophil migration in type IB glycogen storage disease. J Pediatr. 1980. 97:906–910.

10. Kure S, Suzuki Y, Matsubara Y, Sakamoto O, Shintaku H, Isshiki G, Hoshida C, Izumi I, Sakura N, Narisawa K. Molecular analysis of glycogen storage disease type Ib: identification of a prevalent mutation among Japanese patients and assignment of a putative glucose-6-phosphate translocase gene to chromosome 11. Biochem Biophys Res Commun. 1998. 248:426–431.

11. Gerin I, Veiga-da-Cunha M, Noel G, Van Schaftingen E. Structure of the gene mutated in glycogen storage disease type Ib. Gene. 1999. 227:189–195.

12. Hiraiwa H, Pan CJ, Lin B, Moses SW, Chou JY. Inactivation of the glucose 6-phosphate transporter causes glycogen storage disease type 1b. J Biol Chem. 1999. 274:5532–5536.

13. Janecke AR, Bosshard NU, Mayatepek E, Schulze A, Gitzelmann R, Burchell A, Bartram CR, Janssen B. Molecular diagnosis of type 1c glycogen storage disease. Human Genet. 1999. 104:275–277.

14. Chen LY, Pan CJ, Shieh JJ, Chou JY. Structure-function analysis of the glucose-6 phosphate transporter deficient in glycogen storage disease type Ib. Hum Mol Genet. 2002. 11:3199–3207.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download