Abstract
The pathogenic mechanism of ASA-induced urticaria/angioedema (AIU) is still poorly understood, but it has been known that histamine releasing by cutaneous mast cell activation is considered to be an important role. Considering the importance of histamine in AIU, we speculated that a genetic abnormality of histamine-related genes such as a high-affinity IgE receptor, a metabolic enzyme of histamines and histamine receptors, may be involved in the development of AIU. Enrolled in the study were 110 patients with AIU, 53 patients without ASA hypersensitivity who had various drug allergies presenting as exanthematous skin symptoms, and 99 normal healthy controls (NC). Eleven single nucleotide polymorphisms (SNPs) of the β chain of the high-affinity IgE receptor (FCER1B) and three histamine-related genes-histamine N-methyltransferase (HNMT), histamine H1 receptor (HRH1), histamine H2 receptor (HRH2)-were screened using the SNP-IT assay based on a single base extension method. No significant differences were observed in allele and genotype frequencies, and haplotype frequencies of all the SNPs of FCER1B, HNMT, HRH1, and HRH2 among the three groups (p>0.05, respectively). These results suggest that the polymorphisms of FCER1B and the three histamine-related genes may not contribute to the development of AIU phenotype in the Korean population.
ASA (acetylsalicylic acid, or aspirin) hypersensitivity is usually classified into two clinical types: asthma and urticaria with or without angioedema. Although the two manifestations are combined in some patients, ASA-induced urticaria/angioedema (AIU) may affect a different subpopulation from ASA-intolerant asthma (AIA) (1). The prevalence of AIU is approximately 0.3% of the normal population (2) and urticaria symptoms can be aggravated by ASA and other nonsteroidal anti-inflammatory drugs (NSAIDs) in one third of patients with chronic urticaria (3). It has been reported that urinary methylhistamine is increased in chronic urticaria patients with ASA hypersensitivity after ASA provocation test (4). The mechanism of ASA hypersensitivity in cutaneous reaction remain largely unknown, but the key cell in urticaria is mast cell, histamine is a key mediator in the urticaria and its clinical symptoms (erythma, angioedema, itching) can be controlled by antihistamines.
Considering the pathogenic mechanisms of AIU, we can speculate that the polymorphisms of candidate genes involved in histamine release and metabolism and its receptors (H1 and H2 receptors) may be associated with the AIU phenotype. To release histamine from mast cells, the key step is a cross-linking of allergens with IgE, which is bound with the high-affinity IgE receptor (FCER1B) located on their surface (5). Histamine release via autoantibodies against FCER1B was noted in the pathogenic mechanism of chronic idiopathic urticaria (6, 7). Several investigators demonstrated a positive association of gene polymorphism of the β chain of FCER1B with high serum total IgE level, atopy or the asthma phenotype (8-11). Histamine is metabolized primarily by histamine N-methyltransferase (HNMT) and diamine oxidase (5). In addition, the effects of histamine are mediated through two types of receptors: the histamine H1 receptor (HRH1) and the histamine H2 receptor (HRH2). HNMT 314C>T, resulting in a T105I amino acid change, has been shown to induce a functional difference in enzyme activity (12).
In this study, we analyzed eleven known single nucleotide polymorphisms (SNPs) of the FCER1B, HNMT, HRH1, HRH2 and their haplotypes in AIU patients, compared with other drug allergy patients presenting as exanthematous skin symptoms, and in normal healthy controls in a Korean population.
110 patients with AIU, 53 patients having no ASA hypersensitivity with various drug allergies presenting as skin manifestations (especially maculopapular exanthematus rash), and 99 normal healthy controls were enrolled by the Department of Allergy and Rheumatology, Ajou University Hospital, located in the Republic of Korea. All the subjects were Korean. The phenotype of AIU was defined as those having positive results on an ASA oral provocation test, which was performed with 500 mg of ASA (Rhonal®, KunWha Pharmaceutical Co., Seoul, Korea). Patients having ASA intolerant asthma and AIU were excluded. Normal controls, who had non-atopy, no personal and family history of allergic diseases, and no past history of ASA and other drug hypersensitivity, were recruited from the general population. All subjects gave informed consent, which was then approved by the local ethical committee. Skin prick tests were performed with 12 common aeroallergens (Bencard Co., Bredford, U.K.). Atopy was defined as one or more positive reactions to the skin prick test. Chronic urticaria was defined as the daily or near-daily presence of itchy wheals for more than 6 weeks.
Single nucleotide polymorphism (SNP) genotyping was performed by SNP-IT™ assays using the SNPstream 25K™ System (Orchid Biosciences, New Jersey, U.S.A.). Briefly, the genomic DNA region spanning the polymorphic site was amplified using one phosphothiolated primer and one regular PCR primer (Table 1). The amplified PCR products were digested with exonucleases. The 5' phosphthiolates protected one strand of the PCR product from exonuclease digestion, resulting in the generation of a single-stranded PCR template. The single-stranded PCR template was overlaid onto a 384 well plate containing a covalently attached SNP-IT™ primer extension primer designed to hybridize immediately adjacent to the polymorphic site. The SNP-IT™ primer was extended for a single base with a DNA polymerase and mixture of an appropriate acycloterminator, which was labeled with either FITC or biotin and complementary to the polymorphic nucleotide. The identity of the incorporated nucleotide was determined with serial colorimetric reactions with anti-FITC-AP and streptavidin-HRP, respectively. The results of yellow and/or blue color developments were analyzed with the ELISA reader and the final genotype calls were made with the QCReview™ program.
The differences of clinical characteristics between two groups were determined by Student's t-test for continuous variables and chi-square test for categorical variables. The Hardy-Weinberg equilibrium was estimated by chi-square tests. The haplotypes of FCER1B and HNMT were analyzed with the Haplotyper program based on the Bayesian algorithm (13), and linkage disequilibrium between loci were measured using Lewontin's ∣D'∣ (14). Logistic regression models were used for analysis of allele and haplotype frequencies controlling age, sex and atopy as co-variables with alternative models (co-dominant, dominant and recessive models). P values were corrected for multiple comparisons with the pairs of groups of interest using the Bonferroni method (15). A p value of 0.05 or less was regarded as significant. All statistical analyses were performed using the software SPSS, version 10.0 (Chicago, IL, U.S.A.).
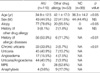
The clinical characteristics of the study subjects are summarized in Table 2. Significant differences existed in the mean age between the patients with other drug allergies and the other two groups (p<0.05, respectively). The prevalence of atopy was 78.6% in AIU patients and 55.5% in patients with other drug allergies, indicating a significant difference (p<0.05). Furthermore, significant differences were found in the prevalence of allergic diseases (including bronchial asthma, allergic rhinitis, allergic conjunctivitis, atopic dermatitis and food allergies) between AIU patients (50.9%) and patients with other drug allergies (11.3%, p<0.01). Thirty three of AIU patients (33.8%) had chronic urticaria.
Genotype distributions of all loci were in the Hardy-Weinberg equilibrium (p>0.05). Allele and genotype frequencies of each SNP of the four candidate genes are shown in Table 3. The frequency of variant allele was extremely low in FCER1B I181L (q=0), HRH1 D349N (q=0) and HRH2 826C>T (q=0.003), being excluded from the statistical analysis. No significant differences were found in allele and genotype frequencies among the three groups with alternative models (codominant, dominant and recessive models). When clinical parameters were analyzed according to the genotypes, serum total IgE levels in AIU patients with FCER1B G237 allele (n=22, 176.2±239.4 IU/mL) was significantly lower than those with the E237 variant (n=79, 374.1±375.0 IU/mL, p=0.004, data not shown). However, no significant differences were observed in atopy rate and serum total IgE level according to the genotypes of the other genes studied (data not shown).
There were four haplotypes in FCER1B having frequencies of more than 1% among the seven haplotypes observed, and three haplotypes in HNMT (Table 4). All SNPs in FCER1B and HNMT showed complete linkage disequilibrium (∣D'∣=1 & r2≠1), and haplotype 1, 2 and 4 of FCER1B are equivalent with FCER1B-109T>C, FCER1B E237G and FCER1B RsaI_ex7 (T>C), respectively, and haplotype 1 and 3 of HNMT are equivalent with HNMT 939A>G and T105I, respectively. No significant differences were found in haplotype frequencies and genotype distributions among the three groups with the alternative model (Table 5).
We investigated 11 SNPs of the four candidate genes (including FCER1B, HNMT, HRH1 and HRH2) that might be associated with AIU pathogenesis. In the present study, no significant differences were found in allele and genotype frequencies of the SNPs and their haplotypes between the study groups, which suggests that these gene polymorphisms may not be related with the development of AIU phenotype in a Korean population.
The high affinity IgE receptor is responsible for initiating allergic response. The binding of an allergen to the receptor-bound IgE leads to mast cell activation and the release of histamines, which are responsible for clinical manifestations of urticaria (16). The receptor is a tetrametric complex composed of an alpha, a beta, and two disulfide-linked gamma chains (17). While the genes for the alpha and gamma subunits are both located on human and mouse chromosome 1 (18), the beta gene is located on 11q13 and spans about 10 kb and contains 7 exons (19). Although polymorphisms of the FCER1B gene has been reported to be associated with atopy, total serum IgE level, bronchial hyperresponsiveness, asthma, and the basophilic histamine-releasing activity of asthmatic patients (8-11, 20), no previous study has been made of the association between FCER1B gene polymorphisms and urticaria/angioedema. This study, therefore, is the first to investigate whether no significant association is found between the FCER1B gene polymorphism with the AIU phenotype in a Korean population. Also, autoimmunity against the high affinity IgE receptor has been reported in chronic urticaria and AIU (6, 7). However, in this study, we could not find any significant associations between the five known SNPs of the FCER1B gene and their haplotypes and AIU phenotype. Further studies in a larger cohort will be needed to confirm these negative results in different ethnic populations.
Histamine is regarded as a leading mediator in acute or chronic urticaria/angioedema and antihistamines are still the main treatment for urticaria/angioedema (5). Histamine is metabolized primarily by HNMT and diamine oxidase in mammals and the effects of histamine are mediated by three pharmacologically defined receptors termed H1 and H2 (5, 21). Large individual variations of HNMT activity is found in human tissues. Preuss et al. (12) have demonstrated that HNMT T105I (C>T) is associated with decreased levels of HNMT enzymatic activity; consequently, the presence of the 314T allele would be expected to result in reduced histamine metabolism. Yan et al. (22) also have shown that the frequency of HNMT 314C>T in asthmatics (q=0.18) is significantly higher than in controls (q=0.14, OR=0.9, p<0.01) in Caucasians. HRH1 mediates the pro-inflammatory actions in the cytokine release and adhesion process (23, 24), while HRH2 suppresses the production of the Th1 inducing cytokine IL-12, resulting in a shift of the Th1/Th2 balance toward Th2 dominance (25). Thus, HRH1 and HRH2 are considered to contribute to the pathogenesis of various allergic diseases. Few studies have been made of these genetic polymorphisms and allergic diseases. Sasaki et al. (26) have shown that no significant association exists between atopic asthma and polymorphisms of HRH1, HRH2, and HNMT genes in a Japanese population. The same SNPs were screened in this study. Although the frequencies of the SNPs were similar with those of the Japanese study, no significant differences were found among the study groups.
In conclusion, these results suggest a lack of associations between FCER1B (-109T>C, RsaI_in2G>A, I181L, E237G, RsaI_ex7T>C), HRH1 (-17C>T, D349A), HRH2 (543G>A, 826C>T), and HNMT (T105I, 939A>G) gene polymorphisms and their haplotypes and the AIU phenotype in a Korean population.
Figures and Tables
References
1. Szczeklik A, Gryglewski RJ, Czerniawska-Mysik G. Clinical patterns of hypersensitivity to nonsteroidal anti-inflammatory drugs and their pathogenesis. J Allergy Clin Immunol. 1977. 60:276–284.

2. Settipane RA, Constantine HP, Settipane GA. Aspirin intolerance and recurrent urticaria in normal adults and children. Epidemiology and review. Allergy. 1980. 35:149–154.
4. Di Lorenzo G, Pacor ML, Vignola AM, Profita M, Esposito-Pellitteri M, Biasi D, Corrocher R, Caruso C. Urinary metabolites of histamine and leukotrienes before and after placebo-controlled challenge with ASA and food additives in chronic urticaria patients. Allergy. 2002. 57:1180–1186.

5. Church MK, Shute JK, Sampson AP. Adkinson NF, Yunginger JW, Busse WW, Bochner BS, Holgate ST, Simons FER, editors. Mast cell-derived mediators. Middleton's allergy principles & practice. 2003. Philadelphia: Mosby, Inc.;189–192.
6. Asero R, Lorini M, Chong SU, Zuberbier T, Tedeschi A. Assessment of histamine-releasing activity of sera from patients with chronic urticaria showing positive autologous skin test on human basophils and mast cells. Clin Exp Allergy. 2004. 34:1111–1114.

7. Lee H, Lee SH, Ro JY, Lee KH. Autoantibody against high affinity IgE receptor in chronic idiopathic urticaria. Korean J Dermatol. 2001. 39:43–49.
8. Hill MR, Cookson WO. A new variant of the β subunit of the high-affinity receptor for immunoglobulin E (Fc ɛRI-β E237G): associations with measures of atopy and bronchial hyperresponsiveness. Hum Mol Genet. 1996. 5:959–962.
9. Hizawa N, Yamaguchi E, Jinushi E, Kawakami Y. A common FCER1B gene promoter polymorphism influences total serum IgE levels in a Japanese population. Am J Respir Crit Care Med. 2003. 161:906–909.
10. Palmer LJ, Paré PD, Faux JA, Moffatt MF, Daniels SE, LeSouëf PN, Bremner PR, Mockford E, Gracey M, Spargo R, Musk AW, Cookson WO. Fc ɛRI-β polymorphism and total serum IgE levels in endemically parasitized Australian aborigines. Am J Hum Genet. 1997. 61:182–188.
11. Shirakawa T, Li A, Dubowitz M, Dekker JW, Shaw AE, Faux JA, Ra C, Cookson WO, Hopkins JM. Association between atopy and variants of the β subunit of the high-affinity immunoglobulin E receptor. Nat Genetics. 1994. 7:125–129.

12. Preuss CV, Wood TC, Szumlanski CL, Raftogianis RB, Otterness DM, Girard B, Scott MC, Weinshilboum RM. Human histamine N-methyltransferase pharmacogenetics: common genetic polymorphisms that alter activity. Mol Pharmacol. 1998. 53:708–717.

13. Niu T, Qin ZS, Xu X, Liu JS. Bayesian haplotype inference for mutiple linked single-nucleotide polymorphisms. Am J Hum Genet. 2002. 70:157–169.
15. Altman DG. Practical statistics for medical research. 1997. London: Chapman & Hall.
16. Dombrowicz D, Flamand V, Brigman KK, Koller BH, Kinet JP. Abolition of anaphylaxis by targeted disruption of the high affinity immunoglobulin E recptor α chain gene. Cell. 1993. 75:969–976.
17. Blank U, Ra C, Miller L, White K, Metzger H, Kinet JP. Complete sturucture and expression in transfected cells of high affinity IgE receptor. Nature. 1989. 337:187–189.
18. Hupp K, Siwarski D, Mock BA, Kinet JP. Gene mapping of the three subunits of the high affinity FcR for IgE to mouse chromosomes 1 and 19. J Immunol. 1989. 143:3787–3791.
19. Sandford AJ, Shirakawa T, Moffatt MF, Daniels SE, Ra C, Faux JA, Young RP, Nakamura Y, Lathrop GM, Cookson WO, Hopkin JM. Localisation of atopy and beta subunit of high-affinity IgE receptor (FCER1) on chromosome 11q. Lancet. 1993. 341:332–334.
20. Kim YK, Oh SY, Oh HB, Chun SY, Cho SH, Koh YY, Min KU, Kim YY. Coding single nucleotide polymorphism in the high-affinity immunoglobulin E receptor β chain (Fc ɛRI-β) gene is associated with immunoglobulin E receptor-mediated histamine release from basophils. Clin Exp Allergy. 2002. 32:751–755.
21. Hill SJ, Ganellin CR, Timmerman H, Schwartz JC, Shankley NP, Young JM, Schunack W, Levi R, Haas HL. International union of pharmacology. XIII. Classification of histamine receptors. Pharmacol Rev. 1997. 49:253–278.
22. Yan L, Galinsky RE, Bernstein JA, Liggett SB, Weinshilboum RM. Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000. 10:261–266.

23. Bachert C. Histamine-a major role in allergy? Clin Exp Allergy. 1998. 28:15–19.
24. Banu Y, Watanabe T. Augmentation of antigen receptor-mediated responses by histamine H1 receptor signaling. J Exp Med. 1999. 189:673–682.

25. van der Pouw Kraan TC, Snijders A, Boeije LC, de Groot ER, Alewijnse AE, Leurs R, Aarden LA. Histamine inhibits the production of interleukin-12 through interaction with H2 receptors. J Clin Invest. 1998. 102:1866–1873.

26. Sasaki Y, Ihara K, Ahmed S, Yamawaki K, Kusuhara K, Nakayama H, Nishima S, Hara T. Lack of association between atopic asthma and polymorphisms of the histamine H1 receptor, histamine H2 receptor, and histamine N-methyl transferase genes. Immunogenetics. 2000. 51:238–240.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download