Abstract
The aims of this study were to estimate the incidences of BCR/ABL, MLL, TEL/AML1 rearrangements, and p16 deletions in childhood acute lymphoblastic leukemia (ALL), to identify new abnormalities, and to demonstrate the usefulness of interphase fluorescence in situ hybridization (FISH). We performed G-banding analysis and FISH using probes for BCR/ABL, MLL, TEL/AML1 rearrangements, and p16 deletions on 65 childhood ALL patients diagnosed and uniformly treated at a single hospital. Gene rearrangements were identified in 73.8% of the patients using the combination of G-banding and FISH, while the chromosomal abnormalities were identified in 49.2% using G-banding alone. Gene rearrangements were disclosed by FISH in 24 (72.7%) of 33 patients with normal karyotype or no mitotic cell in G-banding. Among the gene rearrangements detected by FISH, the most common gene rearrangement was p16 deletion (20.3%) and the incidences of others were 14.1% for TEL/AML1, 11.3% for MLL, and 1.8% for BCR/ABL translocations. Infrequent or new aberrations such as AML1 amplification, MLL deletion, ABL deletion, and TEL/AML1 fusion with AML1 deletion were also observed. We established the rough incidences of gene rearrangements in childhood ALL, found new abnormalities and demonstrated the diagnostic capability of interphase FISH to identify cryptic chromosome aberrations.
Acute lymphoblastic leukemia (ALL) is the most common malignancy in childhood, accounting for approximately 25% of all pediatric malignancies. More than 250 children are newly diagnosed ALL every year in Korea. It is well established that the identification of cytogenetic abnormalities is very useful for the prediction of outcome in childhood ALL. For example, t(9;22), 11q23 abnormalities, and hypodiploidy are known to confer a poor prognosis, and t(12;21), hyperdiploidy are associated with a favorable outcome (1). The association between the deletion of 9p21 and poor prognosis was suggested (2-4). However, questions about the relationship between t(12;21) and good prognosis have arisen because several patients with TEL/AML1 fusion showed poorer clinical outcome if other gene rearrangements coexist (5, 6).
Although detection of chromosome aberrations in ALL has been improved by the development of cytogenetic techniques in conventional G-banding analysis, prognostically important structural or numerical chromosome aberrations may frequently go undetected using conventional G-banding alone due to poor chromosome morphology and few malignant metaphases (7, 8). Especially, 9p abnormalities, t(12;21), and some of the 11q23 rearrangements are very difficult, or even impossible, to detect by conventional G-banding analysis. Interphase fluorescence in situ hybridization (FISH) is a rapid and sensitive tool for detecting gene rearrangements, and thus has been frequently used at the time of diagnosis and during follow-up of patients to monitor minimal residual disease.
There have been few Korean studies evaluating the incidences of BCR/ABL, MLL, TEL/AML1, and p16 gene rearrangements commonly found in childhood ALL because FISH has not yet been routinely used at diagnosis of childhood ALL. Therefore, we cannot confirm the results suggesting the existence of geographical variations in the incidence of TEL/AML1 fusion (9, 10).
In the present study, we performed FISH with probes for BCR/ABL, MLL, TEL/AML1 rearrangements, and p16 deletions for each case of childhood ALL. The aims of this study were to estimate the incidences of different genetic subgroups with abnormalities involving the above genes in Korean childhood ALL, to identify new abnormalities, and to demonstrate the usefulness of FISH.
Among patients diagnosed childhood ALL between 1997 and 2002 at the Samsung Medical Center in Seoul, Korea, 65 patients, whose bone marrow cells had been stored at initial diagnosis and were available for cytogenetic analysis, were studied. The male-female ratio was 1.24 and all patients were treated according to the Children's Cancer Study Group (CCG) protocol. The median age at diagnosis was five years (range, 3 months-15 yr). The leukemia immunophenotype was determined by standard immunofluorescence analysis using a panel of monoclonal antibodies. Patients were classified into three risk groups by prognostic factors such as age, sex, white blood cell (WBC) count, immunophenotype, and other findings (Table 1). A institutional ethical committee approved this study.
Cell culture and chromosome preparation were performed according to two different protocols: synchronized and unsynchronized techniques in parallel. Synchronization was accomplished using methotrexate. Standard cytogenetic preparations were made. At least 20 metaphases were analyzed using Giemsa-trypsin staining. Karyotypes were interpreted according to the International System for Cytogenetic Nomenclature (ISCN) (11).
The selected probes were LSI BCR/ABL ES (extra signal) Dual Color Translocation Probe (Vysis Inc., Downers Grove, IL, U.S.A.), LSI TEL/AML1 ES Dual Color Translocation Probe (Vysis), MLL Dual Color Break Apart Rearrangement Probe (Vysis), and LSI p16/centromere enumeration probe (CEP) 9 Dual Color Probe (Vysis). FISH was performed according to the manufacturer's instructions. The image was analyzed using Cytovision (Applied Imaging International Ltd., Newcastle, England). At least 200 interphase nuclei were analyzed for each case, and if needed, the metaphases were also analyzed. Nuclei with ambiguous signals were excluded from analysis.
To determine cut-off values of each gene rearrangement, we analyzed 20 bone marrow specimens from patients without evidence of hematologic malignancies or solid tumors by FISH. Each cut-off was set at mean proportion of cells with rearrangements plus 3× standard deviations. The cut-off values were as follows: 1.5% for BCR/ABL translocation; 1.0% for MLL translocation; 3.4% for MLL deletion; 1.5% for TEL/AML1 translocation; 3.1% for p16 deletion.
Cytogenetic investigations were performed by conventional G-banding analysis for all cases and by interphase FISH using probes to detect BCR/ABL rearrangements (57 cases), TEL/AML1 rearrangements (64 cases), MLL rearrangements (62 cases), and p16 deletions (64 cases). Numerical and/or structural aberrations were identified in 73.8% of all cases by the combination of conventional G-banding and interphase FISH, while abnormalities were detected in 49.2% of the cases using G-banding alone. Gene rearrangements were disclosed by FISH in 24 (72.7%) of 33 patients who showed a normal banded karyotype or no mitotic cell in G-banding. Among 30 cases harboring structural abnormalities identified by FISH, only three cases showed chromosome aberrations suggesting the FISH results in G-banding analysis (two patients with 11q23 abnormalities and one patient with translocation involving 12p12). Twenty-one patients (32.3%) including two who had been classified as low-risk group but later had relapsed, had prognostically unfavorable gene rearrangements that had not been previously detected by G-banding analysis.
The most common gene rearrangement was p16 deletion (20.3%) and the incidences of others were 14.1% for TEL/AML1, 11.3% for MLL, and 1.8% for BCR/ABL translocations (Table 2).
Five cases showed the coexistence of more than two gene rearrangements. Three cases showed p16 deletions in combination with other gene rearrangements: one case had MLL deletion, TEL/AML1 translocation, and p16 hemizygous deletion; one had MLL translocation and p16 hemizygous deletion; and the other had t(8;14) with p16 homozygous deletion. The other two cases had TEL/AML1 fusions accompanied by rearrangements of non-translocated TEL gene. Table 3 describes the patients with gene rearrangements.
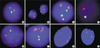
One case displayed only one ABL signal with two BCR signals in 199 of 200 interphase cells (Fig. 1A). The karyotype was 46,XY,del(5)(q32q34). Considering the presence of both signals from the p16 and CEP9 in the p16/CEP9 FISH analysis, this finding indicated deletion of the ABL gene rather than monosomy 9.
Two different types of MLL gene rearrangements were observed in FISH analysis; translocation and deletion. Six of seven cases with MLL translocations showed no chromosome abnormality involving 11q23 in G-banding analysis. Two cases including one with del(11)(q23) in G-banding, showed only one fusion (orange/green) signal in 19% and 41% of 200 interphase cells, respectively (Fig. 1B). Consequently, a large deletion occurring proximal to the MLL breakpoint was suggested. The possibility of monosomy 11 could be excluded because FISH using the CEP11 confirmed the presence of both chromosome 11.
TEL/AML1 translocations were found in nine (14.1%) of 64 patients and none showed t(12;21) in G-banding analysis. Two fusion signals were observed in two cases (Fig. 1C). Two patients (3.1%) with TEL/AML1 fusion had another structural rearrangement involving non-translocated 12p13 breakpoint. One had reciprocal translocation involving 12p12 and 14q21, and the other had deletion of non-translocated TEL gene (Fig. 1D). The proportion of the cells with signals suggesting TEL/AML1 fusion with TEL deletion was 64% of 200 interphase cells. When we performed FISH using the CEP12, 9.0% of 200 cells showed one CEP12 signal and thus we excluded the occurrence of TEL/AML1 fusion in clones with monosomy 12. Four patients showed the loss of one AML1 signal in addition to the TEL/AML1 fusion (Fig. 1E).
One case displayed more than five AML1 signals localized in a very defined chromosomal region in 45% of 200 interphase cells, strongly suggestive of intrachromosomal amplification of the AML1 gene (Fig. 1F).
The combined incidence of hemizygous and/or homozygous deletions determined here was 20.3%. None with p16 deletions had 9p21 abnormalities in G-banding analysis. Homozygous deletions were observed in eight cases (12.5%) and hemizygous deletions in six cases (9.4%). One case had both in two different cell populations. p16 deletions were significantly more common among T-lineage ALL (T-ALL) patients than among precursor-B ALL patients (57.1% vs. 17.2%, p<0.05).
Seven patients were lost on follow-up. All patients except four succeeded in attaining complete remission after induction chemotherapy. Five patients died and seven relapsed. Allogeneic bone marrow transplantations were done in nine patients.
Due to the small number of patients in each genetic subgroup and the short duration of follow-up, we did not analyze the relationship between the outcome and each gene rearrangement. However, eight of fourteen patients with unfavorable outcome showed MLL translocations or p16 deletions.
We performed FISH with probes for BCR/ABL, MLL, TEL/AML1 rearrangements, and p16 deletions to estimate the incidences of different genetic subgroups with abnormalities involving above genes in Korean childhood ALL, to identify new abnormalities, and to demonstrate the usefulness of FISH. A significant increase in detection rate from 49.2% to 73.8% was observed using the combination of conventional G-banding and interphase FISH analysis. Especially of note, FISH was useful to identify the cryptic gene rearrangements in cases with normal banded karyotype or no mitotic cell in G-banding. The conventional G-banding analysis was able to identify the structural abnormalities in few patients with positive FISH results. Moreover, FISH revealed unfavorable gene rearrangements in two patients who had been treated following the protocol for low or intermediate-risk group and eventually relapsed. Therefore, performing FISH at diagnosis would be important to acquire prognostically important information in childhood ALL.
Compared with incidences of other regions, the incidences of TEL/AML1 translocation and p16 homozygous deletion appeared a little lower in our study. The incidence of TEL/AML1 fusion was 14.1% of total childhood ALL and 15.8% of precursor-B ALL in this study. Incidence of around 25% was reported in the United States (12, 13), Germany (14), Italy (14), and France (15). Although differences in technique, criteria for acceptance, and criteria for inclusion in the studies may account for many of these variations, our results also supported the existence of geographical differences in genetic propensity for TEL/AML1 fusion in childhood ALL (9, 10, 16). The incidence of p16 homozygous deletion was 42.9% in T-ALL and 8.6% in precursor-B ALL, lower than that reported (64% in T-ALL and 23% in precursor-B ALL) (17). However, the higher frequency in T-ALL was concordant with previous report (17).
Frequent rearrangements of non-translocated TEL gene were observed in more than 50% of the patients with TEL/AML1 fusion (8, 15, 18), which supports the theory that the TEL/AML1 fusion gene acts in a recessive manner with regard to TEL gene, or that the secondary genetic changes including rearrangements of non-translocated TEL gene are needed in leukemogenesis by TEL/AML1 fusion (19, 20). However, only two (22.2%) cases among nine with TEL/AML1 fusion showed simultaneous rearrangements of non-translocated TEL gene in our study. This may be due to the relatively low sensitivity of the FISH method. By using molecular genetic methods such as RT-PCR, loss of heterozygosity (LOH) analysis, and spectral karyotyping (SKY), the detection of nontranslocated TEL gene rearrangements will be increased. The design of new FISH probes similar to the MLL break-apart probe could be proposed for simultaneous detection of both the translocation and deletion of non-translocated TEL gene. One patient with non-translocated TEL deletion was classified into the high-risk group, whereas other patients with TEL/AML1 fusion alone were classified into low- or intermediate-risk group in our study. Although the clinical significance of TEL deletion is unclear, this finding suggests a certain role for TEL deletion in the progression of the disease (5, 21). Therefore, investigation for the rearrangements of non-translocated TEL gene in patients with TEL/AML1 fusion will be helpful for predicting the prognosis.
Among nine patients with TEL/AML1 fusions, two showed double fusion signals. One displayed trisomy 21 in G-banding analysis and we assumed that the additional chromosome 21 might be not the normal chromosome but the der(21) t(12;21). It was also reported that the additional fusion signals resulted from duplication of der(21)t(12;21) or ider(21) (q10)t(12;21) (10). Because the TEL/AML1 fusion transcript encodes a strong repressor that interferes with AML1-dependent transcription activation and the wild-type TEL gene, the two fusion transcripts may result in increased expression of the TEL/AML1 fusion gene and increased silencing of the wild-type TEL gene (19, 22). In support of this hypothesis, extra copies of der(21)t(12;21) were found more frequently among patients suffering from relapse (23); our case had deletion of non-translocated TEL.
A large proportion of presumptive del(11)(q23) or del(11) (q23q25) might represent previously unidentified translocations that could be detected by FISH (24, 25). However, our patient with del(11)(q23) in G-banding analysis had also deletion of the MLL gene in FISH. Our two patients with MLL deletions showed relatively longer survival, which was in concordance with the report that MLL deletion was associated with good prognosis (25). As different prognoses between the patients with MLL translocation and those with MLL deletion were reported, FISH would be needed in patients with del(11)(q23) in G-banding analysis.
New findings observed in this study were the deletions of the ABL and AML1 gene. Deletion of ABL was observed in a 13-yr-old boy with T-ALL. Deletions of the 9q34 region on which the ABL gene is located have been known to be quite common findings in several solid tumors (26, 27). However, the deletions of 9q34 have rarely been found in ALL and thus the clinical significance is unknown. Searching for identical cases and additional follow-up study of our case will be helpful in understanding the role of ABL deletion in leukemogenesis of ALL. As well, deletion of AML1 was accompanied with TEL/AML1 fusion and observed in four female patients with precursor-B ALL, which has not yet been reported. It was unclear whether the deletion occurred at der(21) or at normal chromosome 21. The loss of 21q was not observed in patients with TEL/AML1 fusion, while the gain of 21q was frequently found (19). As such, we assumed that the deletions might occur at the der(21) and the AML1 deletion would be one of the secondary genetic changes required in leukemogenesis by TEL/AML1 fusion.
Amplification of the AML1 gene was observed in a 10-yr-old male patient with precursor-B ALL. Since first reported (28), ten patients with AML1 amplification have been subsequently reported (7, 29-32). All cases had childhood precursor-B ALL. One of ten showed TEL/AML1 fusion and none showed TEL deletion. Although six cases were initially classified into the high-risk group, all but one remained alive without relapse. Our case also remained alive without events. AML1 amplification was suggested that play an important role in leukemogenesis as a target event in a trisomy 21 or a 21q22 amplicon (31). Further studies are needed to know the role of amplification of the AML1 gene in leukemogenesis of ALL.
The frequent presence of TEL and p16 rearrangements in the company of other genetic changes suggested that the rearrangements of tumor suppressor genes might contribute to leukemogenesis in cooperation with other genetic changes, possibly by amplifying the malignant potential.
In conclusion, routine performance of interphase FISH using BCR/ABL, MLL, TEL/AML1, and p16 probes at diagnosis would be very useful to establish accurate prognosis and to monitor the minimal residual disease in childhood ALL. Further study with a larger number of patients would be necessary to know the relationship between the outcome and each gene rearrangement and to provide a better understanding of leukemogenesis by new gene rearrangements identified in this study.
Figures and Tables
Fig. 1
Abnormal signal patterns in FISH analysis. (A) One ABL gene locus is deleted and both BCR genes are present as indicated by one orange and two green signals (1O2G), respectively. (B) Only one fusion signal indicating deletion of one MLL gene. (C) Two TEL/AML1 fusion signals (2O1G2F) indicating double fusion genes. (D) A TEL/AML1 fusion with TEL deletion (2O1F). (E) TEL/AML1 fusion with AML1 deletion (1O1G1F). (F) More than five orange signals indicating AML1 gene amplification are present. (G) A nucleus containing p16 hemizygous deletion showing one orange and two green signals (1O2G). (H) A nucleus containing p16 homozygous deletion showing two green signals without orange signals (2G).
References
1. Ma SK, Wan TS, Chan LC. Cytogenetics and molecular genetics of childhood leukemia. Hematol Oncol. 1999. 17:91–105.

2. Kees UR, Burton PR, Lü C, Baker DL. Homozygous deletion of the p16/MTS1 gene in pediatric acute lymphoblastic leukemia is associated with unfavorable clinical outcome. Blood. 1997. 89:4161–4166.

3. Heerema NA, Sather HN, Sensel MG, Liu-Mares W, Lange BJ, Bostrom BC, Nachman JB, Steinherz PG, Hutchinson R, Gaynon PS, Arthur DC, Uckun FM. Association of chromosome arm 9p abnormalities with adverse risk in childhood acute lymphoblastic leukemia: a report from the children's cancer group. Blood. 1999. 94:1537–1544.
4. Carter TL, Watt PM, Kumar R, Burton PR, Reaman GH, Sather HN, Baker DL, Kees UR. Hemizygous p16INK4A deletion in pediatric acute lymphoblastic leukemia predicts independent risk of relapse. Blood. 2001. 97:572–574.
5. Wiemels JL, Ford AM, Van Wering ER, Postma A, Greaves M. Protracted and variable latency of acute lymphoblastic leukemia after TEL-AML1 gene fusion in utero. Blood. 1999. 94:1057–1062.
6. Anguita E, Gonzalez FA, Lopez J, Villegas A. TEL/AML1 transcript and p16 gene deletion in a patient with childhood acute lymphoblastic leukemia. Br J Haematol. 1997. 99:240–241.
7. Nordgren A, Schoumans J, Söderhäll S, Nordenskjöld M, Blennow E. Interphase fluorescence in situ hybridization and spectral karyotyping reveals hidden genetic aberrations in children with acute lymphoblastic leukemia and a normal banded karyotype. Br J Haematol. 2001. 114:786–793.
8. Nordgren A, Heyman M, Sahlén S, Schoumans J, Söderhäll S, Nordenskjöld M, Blennow E. Spectral karyotyping and interphase FISH reveal abnormalities not detected by conventional G-banding. Eur J Haematol. 2002. 68:31–41.
9. Garcia-Sanz R, Alaejos I, Orfão A, Rasillo A, Chillón MC, Tabernero MD, Mateos MV, López-Pérez R, González D, Balanzategui A, González M, San Miguel JF. Low frequency of the TEL/AML1 fusion gene in acute lymphoblastic leukemia in Spain. Br J Haematol. 1999. 107:667–669.
10. Andreasson P, Höglund M, Békássy AN, Garwicz S, Heldrup J, Mitelman F, Johansson B. Cytogenetic and FISH studies of a single center consecutive series of 152 childhood acute lymphoblastic leukemias. Eur J Haematol. 2000. 65:40–51.

11. Mitelman F, editor. ISCN. An international system for human cytogenetic nomenclature. 1995. Basel: Tennessee, S Karger.
12. McLean TW, Ringold S, Neuberg D, Stegmaier K, Tantravahi R, Ritz J, Koeffler HP, Takeuchi S, Janssen JW, Seriu T, Bartram CR, Sallan SE, Gilliland DG, Golub TR. TEL/AML-1 dimerizes and is associated with a favorable outcome in childhood acute lymphoblastic leukemia. Blood. 1996. 88:4252–4258.

13. Rubnitz JE, Shuster JJ, Land VJ, Link MP, Pullen DJ, Camitta BM, Pui CH, Downing JR, Behm FG. Case-control study suggests a favorable impact of TEL rearrangement in patients with B lineage acute lymphoblastic leukemia treated with antimetabolite based therapy: a Pediatric Oncology Group study. Blood. 1997. 89:1143–1146.
14. Borkhardt A, Cazzaniga G, Viehmann S, Valsecchi MG, Ludwig WD, Burci L, Mangioni S, Schrappe M, Riehm H, Lampert F, Basso G, Masera G, Garbott J, Biondi A. Incidence and clinical relevance of TEL/AML1 fusion genes in children with acute lymphoblastic leukemia enrolled in the German and Italian multicenter therapy trials. Blood. 1997. 90:571–577.

15. Cavé H, Cacheux V, Raynaud S, Brunie G, Bakkus M, Cochaux P, Preudhomme C, Lai JL, Vilmer E, Grandchamp B. ETV6 is the target of chromosome 12p deletions in t(12;21) childhood acute lymphocytic leukemia. Leukemia. 1997. 11:1459–1464.

16. Park KU, She CJ, Shin HY, Ahn HS, Kim CJ, Cho BK, Cho HI, Lee DS. Low incidence of TEL/AML1 fusion and TEL deletion in Korean childhood acute leukemia by extra-signal fluorescence in situ hybridization. Cancer Genet Cytogenet. 2001. 126:73–77.

17. Drexler HG. Review of alterations of the cyclin dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemialymphoma cells. Leukemia. 1998. 12:845–859.
18. Raynaud S, Cavée H, Baens M, Shurtleff SA, Mathew S, Raimondi S. The 12;21 translocation involving TEL and deletion of the other TEL allele: two frequently associated alterations found in childhood acute lymphoblastic leukemia. Blood. 1996. 87:2891–2899.

19. Raynaud SD, Dastugue N, Zoccola D, Shurtleff SA, Mathew S, Raimondi SC. Cytogenetic abnormalities associated with the t(12;21). A collaborative study of 169 children with t(12;21)-positive acute lymphoblastic leukemia. Leukemia. 1999. 13:1325–1330.

20. Kempski HM, Sturt NT. The TEL-AML1 fusion accompanied by loss of the untranslocated TEL allele in B-precursor acute lymphoblastic leukemia of childhood. Leukemia Lymphoma. 2000. 40:39–47.
21. Kim DH, Moldwin RL, Vignon C, Bohlander SK, Suto Y, Giordano L, Gupta R, Fears S, Nucifora G, Rowley JD, Smith SD. TEL-AML1 translocations with TEL and CDKN2 inactivation in acute lymphoblastic leukemia cell lines. Blood. 1996. 88:785–794.

22. Inamdar N, Kumar SA, Banavali SD, Advani S, Magrath I, Bhatia K. Comparative incidence of the rearrangements of TEL/AML1 and ALL1 genes in pediatric precursor B acute lymphoblastic leukemias in India. Int J Oncol. 1998. 13:1319–1322.

23. Loncarevic IF, Roltzheim B, Ritterbach J, Viehmann S, Borkhardt A, Lampert F, Harbott J. Trisomy 21 is a recurrent secondary aberration in childhood acute lymphoblastic leukemia with TEL/AML1 gene fusion. Genes Chromosomes Cancer. 1999. 24:272–277.
24. Kobayashi H, Espinosa R III, Thirman MJ, Fernald AA, Shannon K, Diaz MO, Le Beau MM, Rowley JD. Do terminal deletions of 11q23 exist? Identification of undetected translocations with fluorescence in situ hybridization. Genes Chrom Cancer. 1993. 7:204–208.

25. Harbott J, Mancini M, Verellen-Dumoulin C, Moorman AV, Secker-Walker LM. Hematological malignancies with a deletion of 11q23: cytogenetic and clinical aspects. Leukemia. 1998. 12:823–827.

26. Devlin J, Elder PA, Gabra H, Steel CM, Knowles MA. High frequency of chromosome 9 deletion in ovarian cancer: evidence for three tumour suppressor loci. Br J Cancer. 1996. 73:420–423.
27. Takita J, Hayashi Y, Kohno T, Yamaguchi N, Hanada R, Yamamoto K, Yokota J. Deletion map of chromosome 9 and p16 (CDKN2A) gene alterations in neuroblastoma. Cancer Res. 1997. 57:907–912.
28. Niini T, Kanerva J, Vettenranta K, Saarinen-Pihkalla UM, Knuutila S. AML1 gene amplification: a novel finding in childhood acute lymphoblastic leukemia. Haematologica. 2000. 85:362–366.
29. Cin PD, Atkins L, Ford C, Ariyanayagam S, Armstrong SA, George R, Cleary A, Morton CC. Amplification of AML1 in childhood acute lymphoblastic leukemia. Genes Chrom Cancer. 2001. 30:407–409.
30. Coniat MB, Khac FN, Daniel MT, Bernard OA, Berger R. Chromosome 21 abnormalities with AML1 amplification in acute lymphoblastic leukemia. Genes Chrom Cancer. 2001. 32:244–249.
31. Ma SK, Wan TS, Cheuk AT, Fung LF, Chan GC, Chan SY, Ha SY, Chan LC. Characterization of additional genetic events in childhood acute lymphoblastic leukemia with TEL/AML1 gene fusion: a molecular cytogenetic study. Leukemia. 2001. 15:1442–1447.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download