Abstract
Previous molecular genetic studies of laryngeal squamous cell carcinoma (SCC)have shown certain chromosomal regions with recurring alterations. But studies of sequential molecular alterations and genetic progression model of laryngeal SCC have not been clearly defined. To identify the chromosomal alterations associated with the carcinogenesis of laryngeal SCC, we analyzed genomic DNA from microdissected squamous metaplasia, squamous dysplasia, invasive SCC, and metastatic carcinoma samples from 22 laryngeal SCC patients for loss of heterozygosity (LOH) at microsatellite loci. Ten microsatellite markers on chromosome 3p, 8p, 9p, and 17p were used. LOH at 9p21 was observed in the all stages including squamous metaplasia, squamous dysplasia, invasive SCC and metastatic carcinoma. LOH at 17p13.1, 3p25 and 3p14.2 was observed from the squamous dysplasia, invasive SCC and metastatic carcinoma. LOH at 8p21.3-p22 was observed mainly from the invasive SCC and metastatic carcinoma. The results suggest that 9p21 in the early event, 17p13.1, 3p25 and 3p14.2 in the intermediate event and 8p21.3-p22 in the late event may be involved in the laryngeal carcinogenesis.
With recent development of molecular biology, cancer was revealed as a genetic disease. Unlike other hereditary diseases, cancer is a more complex genetic disease in that cancer does not develop just with one gene abnormality but with many genetic alterations that participate within one cell.
Currently, most of solid tumors are known to occur with the process of genetic alterations being accumulated in stages; Fearon and Vogelstein suggested a multi-step progression model of genetic alterations that participates in the initiation and progression of colon cancer through the research on the tumor progression from dysplastic polyp and adenoma to adenocarcinoma (1).
We think that laryngeal squamous cell carcinoma (SCC) also progresses through well-established clinical and histopathological stages; and there have been many studies on genetic alterations related to laryngeal SCC (2-4). However, it is not clear yet which genes participate in histopathological stages of laryngeal SCC.
Among many genes related to the development of tumors, the putative tumor suppressor gene has the gene code on the protein that suppresses cell growth and thus, is known to suppress tumor formation; and inactivation of this gene plays a definite role in the development and progression of tumors. Loss of heterozygosity (LOH) is the most common phenomenon in the inactivation of the putative tumor suppressor gene; and LOH analyses have provided definite clues to confirming the position of putative tumor suppressor gene and studying the process of cloning (5).
From the paraffin embedded tissues of larynx and cervical lymph node obtained from the same larynx SCC patients, the authors of the present study selectively separated the cells of each stage, i.e. normal, squamous metaplasia, squamous dysplasia, invasive SCC, and metastatic carcinoma of cervical lymph node, using the microdissection technique. We tried to confirm the progressive stages of genetic alterations related to the progression of laryngeal SCC.
Twenty-two cases of laryngeal SCC with cervical lymph node metastasis were selected from the clinical pathology department of The Catholic University of Korea, College of Medicine. All cases were confirmed to contain foci of squamous metaplasia, squamous dysplasia, invasive SCC and metastatic carcinoma in their pathologic materials.
Cells from the areas of squamous metaplasia, squamous dysplasia, invasive SCC and metastatic carcinoma of the cervical lymph nodes were selectively produced from H&E stained slides without normal cell contamination using a 30 G needle (Fig. 1-1, Fig. 1-2). We also obtained normal lymphocytes or submucosal glands for corresponding normal DNA from the same cases.
DNA extraction was done by a modified single step DNA extraction method. Procured cells (500 cells) in 20 µL of DNA extraction buffer containing 100 mM Tris-HCl, pH 8.0; 0.1% Tween 20 and 0.1 mg/mL proteinase K, were incubated at 52℃ for 1 day. The mixture was boiled for 10 min to inactivate the proteinase K and 2 µL of this solution were used as DNA template for PCR amplification.
DNA from each lesions and corresponding normal cells were amplified by thermal cycler with ten microsatellite markers (Research Genetics, Huntsville, AL) including IFNA, D9S171 and D9S104 on 9p21, TP53 on 17p13.1, D3S1038 on 3p25, D3S1067 and D3S1234 on 3p14.2, D8S133, D8S261, D8S258 on 8p21.3-p22. Each PCR reaction was generally performed in standard conditions in a 10 µL of reaction mixture containing 2 µL of template DNA, 0.25 µL each primers, 1.25 mM each dNTP 1 µL, 1.5 mM MgCl2 0.5 µL, 10×PCR buffer 1 µL, 0.4 U Taq polymerase 0.07 µL (Perkin-Elmer, Foster City, CA), and 10 mCi [32P] CTP 0.05 µL. The reaction mixtures were denatured at 95℃ and incubated 32 cycles (denaturing at 94℃ for 40 sec, annealing at 55℃ for 90 sec and extending 72℃ for 90 sec) with some variations made in the annealing temperature. Reaction products (8 µL) were denatured and electrophoresed in 6% polyacrylamide gels containing 7 M urea. After electrophoresis, the gels were transferred to 3 mm Whatman paper then dried, and finally autoradiography was performed using Kodak-OMAT film (Eastman Kodak, Ochester, NY).
The results of LOH according to the microsatellite markers selected for the present study and histopathological stages of each sample are shown in Table 1. The status of allelic loss was compared between the normal cell, squamous metaplasia, squamous dysplasia, invasive SCC, and metastatic carcinoma of lymph node from the same patients. LOH in case 2 and case 13 are shown in Fig. 2 and Fig. 3, respectively.
LOH on IFNA, D9S171 and D9S104 was detected in 63.6% (7/11), 57.1% (4/7) and 50.0% (4/8), respectively. Most of these cases except three cases showed LOH in all histopathologic stages, from squamous metaplasia to metastatic carcinoma. Two cases (case 1 in INFA and case 21 in D9S171) among three cases showed LOH in squamous dysplasia, invasive and metastatic carcinoma. Another case (case 12 in IFNA) showed LOH in only invasive and metastatic carcinoma. From these results, we guess the genetic alterations of 9p21 participate when the normal cells transform to squamous metaplastic cells. Since p16ink4a gene is located between IFNA and D9S171, a LOH on either of the two markers indicates a genetic alteration on p16ink4a gene. And the presence of another tumor suppressor gene around D9S104 is suggested.
LOH on TP53 was detected in 7 (41.2%) of 17 informative cases from squamous dysplasia, invasive carcinoma and metastatic carcinoma. There was no LOH in all 17 informative cases of squamous metaplasia. Thus, we guess that the genetic alterations of 17p13.1 participate in the transformation from non-neoplastic stage to preinvasive neoplastic stage.
D3S1038, on 3p25, showed no LOH in all 10 cases of squamous metaplasia, showed LOH in 6 (60.0%) of 10 informative cases of squamous dysplasia, invasive carcinoma and metastatic carcinoma. Thus, we guess that the genetic alterations of 3p25 participate in the transformation to preinvasive neoplastic lesion.
Also in D3S1067 and D3S1234 on 3p14.2, LOH was detected in 57.1% (8/14) and 66.7% (10/15), respectively. All cases except one case (case 18 in D3S1067) showed LOH in squamous dysplasia, invasive carcinoma and metastatic carcinoma. In case 18, LOH was not present in squamous dysplasia, but observed in only invasive and metastatic carcinoma. Thus, we guess that the genetic alterations of 3p14.2 participate in the transformation to preinvasive neoplastic stage.
LOH on D8S133 was detected in 6 (54.5%) of 11 informative cases. In all 11 informative cases, there was no LOH in squamous metaplasia. Among these 6 cases, 3 cases showed LOH in squamous dysplasia, invasive carcinoma and metastatic carcinoma. Remaining 3 cases showed LOH in only invasive carcinoma and metastatic carcinoma.
LOH on D8S261 was observed in 8 (88.9%) of 9 informative cases, and 4 of these 8 cases showed LOH in invasive carcinoma and metastatic carcinoma, and not in squamous dysplasia. In the other 4 cases, LOH was observed in the squamous dysplasia, invasive carcinoma and metastatic carcinoma.
D8S258 also showed no LOH in squamous metaplasia of all 10 informative cases. 5 (50.0%) of 10 cases showed LOH, with 4 cases showing LOH in invasive carcinoma and metastatic carcinoma. In the other case, LOH was shown in squamous dysplasia, invasive carcinoma and metastatic carcinoma. Thus, we guess that the tumor suppressor gene is presumed to be on 8p21.3-p22 and may participate mainly in the transition stage from preinvasive neoplastic stage to invasive carcinoma. Since the frequency of LOH was the highest on D8S261 among the 3 markers, we believe that there is a possibility that the related gene is present near D8S261 (8p22).
In the past several years, by comparing and relating the genetic alterations and histopathological progression in several types of tumors, there have been studies on coming up with a model of tumor development and progression. After Fearon and Vogelstein (1) first described the molecular genetic progression model of colon cancer, the models of tumor development and progression on brain tumor and urinary bladder cancer were also studied (4, 5). Since then, Califarno et al. have proposed a preliminary model on the progression of head and neck SCC (6). The chromosomes 9p, 17p, 3p and 8p are where gene losses occur most frequently in head and neck SCC. The chromosome 9p21 (7, 8) includes p16INK4a which encodes the p16, a cyclin dependent kinase inhibitor. 17p13.1 (9, 10) is known as where p53 gene is located on. The chromosome 3p is where the tumor suppressor gene for HNSCC is presumed to be located on at least 3 sites (3p25, 3p21, 3p14) (11, 12), and among these 3, there are VHL gene (3p25) related to von Hippel-Lindau disease (13) and Fragile Histidine Triad (FHIT) gene (3p14.2) (14). Although the location of the tumor suppressor gene on the chromosome 8p is not clear, Fujiwara et al. (15) suggested that the site 8p21.3-p22 is where the tumor suppressor gene might be present since it is the site of frequent damages in hepatocellular carcinoma, colon cancer, and non-small cell lung cancer.
The authors of the present study selectively collected the cells from each histopathological stage of laryngeal SCC, i.e., normal, squamous metaplasia, squamous dysplasia, invasive SCC, and metastatic carcinoma of cervical lymph node, in the same patients and tried to observe a series of sequential genetic alterations in these cells. Also, by limiting the study population to only those with laryngeal SCC, we tried to eliminate possible different effects of different anatomical sites.
LOH of 9p21 was observed from the squamous metaplasia stage in the majority cases. Thus, we think that the genetic alterations of 9p21 participate in the early stage of tumorigenesis where normal cells change to squamous metaplasia stage. p16INK4a, which is located between IFNA and D9S171 or another tumor suppressor gene around the D9S104 might be related with squamous metaplasia. This LOH of 9p21 is also reported in many other solid tumors, such as urinary bladder cancer, malignant melanoma, lung cancer, and brain tumor (16-19), and is reported to play a role in the early stage of tumor development (18, 20).
LOH on TP53 was shown in 41.2% (7/17). In these cases, LOH was present in squamous dysplasia, invasive carcinoma and metastatic carcinoma; thus, we think that the genetic alteration of 17p13.1 plays a role in the transformation to preinvasive neoplastic stage.
As 3p25 and 3p14.2, LOH was observed in the squamous dysplasia, invasive carcinoma and metastatic carcinoma but not in the squamous metaplasia. Thus, as same as 17p13.1, we think that the genetic alterations of 3p25 and 3p14.2 participate in transformation to preinvasive neoplastic stages.
Since the 8p21.3-p22 site is the most frequently damaged site in hepatocellular carcinoma, colon cancer, and non-small cell lung carcinoma, it is presumed that at least one tumor suppressor gene is present on this site (21, 22); and although Fujiwara et al. (15) mentioned about a tumor suppressor gene called PDGF receptor beta like tumor suppressor gene, the amino acid sequence for the gene has not yet known. The markers for this site, D8S133, D8S261, and D8S258, also did not show LOH in squamous metaplasia areas. Some cases showed LOH in squamous dysplasia and other cases in invasive carcinoma. All these markers showed highly frequent LOH, showing a possible tumor suppressor gene related to SCC at the 8p21.3-p22, and we think that this gene might participate in the late stage of tumor development, from squamous dysplasia to invasive carcinoma. Among the 3 markers, D8S261 showed especially high frequency of LOH, showing a possibility that the related gene might be present near D8S261. Scholnick et al. (23) reported that genetic alterations of 8p are related with recurrence or bad prognosis of larynx cancer.
Among the studies on the relationship between genetic alterations and lymph node metastasis of head and neck SCC, Ogawara et al. (24) reported that the allelic loss of the chromosome 13q14.3 is related with cervical lymph node metastasis in SCC of oral cavity, whereas Scholnick et al. (23) reported that the chromosomal alterations of 3p, 5q, 8p, 9p, 9q, 13q, and 17p in supraglottic SCC do not have a significant relationship with lymph node metastasis. Recent study reported evidences for the putative role of RB1 gene alteration in metastatic process of laryngeal SCC (25). In the present study, we collected primary carcinoma and metastatic carcinoma tissue from the same patient, and observed and compared LOH. In all cases, both primary and metastatic carcinomas manifested identical patterns of LOH. We think that another genetic factor might participate in the process of metastasis of invasive carcinoma.
In laryngeal SCC, clarifying the genetic progression is necessary to understand the development, behavior and progression of laryngeal SCC. Knowledge on early genetic alterations can help the early detection and screening of tumors, and at the time of tumor resection, by analyzing the molecular biological state of the cells around the resected area, a more precise evaluation of the success of resection can be achieved (26). Also, deciding the molecular biological state in benign mucosal epithelium around the invasive carcinoma is important in predicting whether this area might transform into more malignant, i.e. invasive carcinoma stage. Veltman et al. (27) reported chromosomal tetraploidization as an indicator of malignant progression in laryngeal mucosa. At the same time, knowledge on genetic alterations of tumor progression can make a new tumor classification possible by adding a molecular biological information and may become the basis for gene therapy in the future. In the present study, we suggest that 9p21 in the early event, 17p13.1, 3p25 and 3p14.2 in the intermediate event and 8p21.3-p22 in the late event may be involved in the laryngeal carcinogenesis.
Figures and Tables
Fig. 1-1
Sections of laryngeal carcinoma (A) and metastatic lymph node (B) from case 4 showing the areas of metaplasia (M), dysplasia (D), invasive carcinoma (I), and lymph node metastatic carcinoma (L) (H&E, ×4).

Fig. 1-2
The magnified areas of metaplasia (M), dysplasia (D), invasive carcinoma (I), and lymph node metastatic carcinoma (L) before (A) and after (B) microdissection (H&E, ×100).
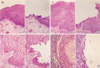
Fig. 2
Analysis of loss of heterozygosity according to the development and progression of laryngeal squamous cell carcinoma (case 2) showing allelic loss (arrow head) from metaplasia (IFNA, D9S171, D9S104) and from dysplasia (TP53, D3S1038, D3S1067), Four markers (D3S1234, D8S261, D8S258, D8S133) are homozygote (non-informative).
N, normal; M, metaplasia; D, dysplasia; I, invasive carcinoma; L, lymph node metastasis.
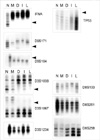
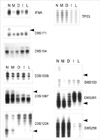
Fig. 3
Analysis of loss of heterozygosity according to the development and progression of laryngeal squamous cell carcinoma (case 13) showing allelic loss (arrow head) from metaplasia (D9S171), from dysplasia (D3S1067, D3S1234, D8S133), and from invasive carcinoma (D8S261, D8S258). Four markers (IFNA, D9S104, TP53, D3S1038) are homozygote (non-informative).
N, normal; M, metaplasia; D, dysplasia; I, invasive carcinoma; L, lymph node metastasis.
References
2. Ah-See KW, Cooke TG, Pickford IR, Soutar D, Balmain A. An allelotype of squamous carcinoma of the head and neck using microsatellite markers. Cancer Res. 1994. 54:1617–1621.
3. Nawroz H, van der Riet P, Hruban RH, Koch W, Ruppert JM, Sidransky D. Allelotype of head and neck squamous cell carcinoma. Cancer Res. 1994. 54:1152–1155.
4. Sidransky D, Mikkelsen T, Schwechheimer K, Rosenblum ML, Cavanee W, Vogelstein B. Clonal expansion of p53 mutant cells is associated with brain tumor progression. Nature. 1992. 355:846–847.
5. Simoneau AR, Jones PA. Bladder cancer: the molecular progression to invasive disease. World J Urol. 1994. 12:89–95.

6. Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D. Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 1996. 56:2488–2492.
7. Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS 3rd, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994. 264:436–440.

8. Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994. 368:753–756.

9. Boyle JO, Hakim J, Koch W, van der Riet P, Hruban RH, Roa RA, Correo R, Eby YJ, Ruppert JM, Sidransky D. The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res. 1993. 53:4477–4480.
10. Soder AI, Hopman AH, Ramaekers FC, Conradt C, Bosch FX. Distinct nonrandom patterns of chromosomal aberrations in the progression of squamous cell carcinomas of the head and neck. Cancer Res. 1995. 55:5030–5037.
11. Maestro R, Gasparotto D, Vukosavljevic T, Barzan L, Sulfaro S, Boiocchi M. Three discrete regions of deletion at 3p in head and neck cancers. Cancer Res. 1993. 53:5775–5779.
12. Wu CL, Sloan P, Read AP, Harris R, Thakker N. Deletion mapping on the short arm of chromosome 3 in squamous cell carcinoma of the oral cavity. Cancer Res. 1994. 54:6484–6488.
13. Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, Schmidt L, Zhou F, Li H, Wei MH, Chen F, Glenn G, Choyke P, Walther MU, Weng Y, Duan DSR, Dean M, Glavac D, Richards FM, Crossey PA, Ferguson-Smith MA, Le Pastier D, Chumakov I, Cohen D, Chinault C, Maher ER, Linehan WM, Zbar B, Lerman MI. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993. 260:1317–1320.

14. Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996. 84:587–597.

15. Fujiwara Y, Ohata H, Kuroki T, Koyama K, Tsuchiya E, Monden M, Nakamura Y. Isolation of a candidate tumor suppressor gene on chromosome 8p21.3-p22 that is homologous to an extracellular domain of the PDGF receptor beta gene. Oncogene. 1995. 10:891–895.
16. Coleman A, Fountain JW, Nobori T, Olopade OI, Robertson G, Housman DE, Lugo TG. Distinct deletions of chromosome 9p associated with melanoma versus glioma, lung cancer and leukemia. Cancer Res. 1994. 54:344–348.
17. Devlin J, Keen AJ, Knowles MA. Homozygous deletion mapping at 9p21 in bladder carcinoma defines a critical region within 2cM of IFNA. Oncogene. 1994. 9:2757–2760.
18. Isshiki K, Seng BA, Elder DE, Guerry D, Linnenbach AJ. Chromosome 9 deletion in sporadic and familial melanomas in vivo. Oncogene. 1994. 9:1649–1653.
19. Merlo A, Gabrielson E, Mabry M, Vollmer R, Baylin SB, Sidransky D. Homozygous deletion on chromosome 9p and loss of heterozygosity on 9q, 6p, and 6q in primary human small cell lung cancer. Cancer Res. 1994. 54:2322–2326.
20. Nees M, Homann N, Discher H, Andl T, Enders C, Herold-Mende C, Schuhmann A, Bosch FX. Expression of mutated p53 occurs in tumor-distant epithelia of head and neck cancer patients: a possible molecular basis for the development of multiple tumors. Cancer Res. 1993. 53:4189–4196.
21. Emi M, Fujiwara Y, Nakajima T, Tsuchiya E, Tsuda H, Hirohashi S, Maeda Y, Tsuruta K, Miyaki M, Nakamura Y. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Res. 1992. 52:5368–5372.
22. Chinen K, Isomura M, Izawa K, Fujiwara Y, Ohata H, Iwamasa T, Nakamura Y. Isolation of 45 exon-like fragments from 8p22→p21.3, a region that is commonly deleted in hepatocellular, colorectal, and non-small cell lung carcinomas. Cytogenet Cell Genet. 1996. 75:190–196.
23. Scholnick SB, El-Mofty SK, Shaw ME, Sunwoo JB, Haughey BH, Sun PC, Piccirillo JF, Zequeira MR. Clinical correlations with allelotype in supraglottic squamous cancer. Otolaryngol Head Neck Surg. 1998. 118:363–370.

24. Ogawara K, Miyakawa A, Shiba M, Uzawa K, Watanabe T, Wang XL, Sato T, Kubosawa H, Kondo Y, Tanzawa H. Allelic loss of chromosome 13q14.3 in human oral cancer: Correlation with lymph node metastasis. Int J Cancer. 1998. 79:312–317.

25. Kujawski M, Rydzanicz M, Sarlomo-Rikala M, Szyfter K. Rearrangements involving the 13q chromosome arm committed to the progression of laryngeal squamous cell carcinoma. Cancer Genet Cytogenet. 2002. 137:54–58.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download