Abstract
To gain molecular understanding of carcinogenesis of breast cancer, gene expression profiles were analyzed using cDNA microarray representing 4,600 cDNAs in 10 breast cancer samples and the adjacent noncancerous breast tissues from the same patients. The alterations in gene expression levels were confirmed by reverse-transcription PCR in four randomly selected genes. Genes that were differently expressed in cancer and noncancerous tissues were identified. 106 (of which 55 were known) and 49 (of which 28 were known) genes were up- or down-regulated, respectively, in greater than 60% of the breast cancer samples. In cancer tissues, genes related to cell cycle, transcription, metabolism, cell structure/motility and signal transduction were mostly up-regulated. Furthermore, three cancer tissues showing immunohistochemically aberrant accumulation of β-catenin in the nucleus and/or cytoplasm revealed down-regulation of Siah and Axin genes and up-regulation of Wnt and c-myc genes. These findings were highly consistent with Wnt signaling pathway associated with β-catenin regulation previously suggested by others. Our studies, therefore, provide not only a molecular basis to understand biological processes of breast cancer but also useful resources to define the mechanism of β-catenin expression in tumorigenesis of breast cancer.
Recently, there has been an increase in the numbers of breast cancer diagnosed in Korea. Breast cancer patients with the same stage of disease can have markedly different treatment responses and overall outcome. Furthermore, the biology of breast cancer remains poorly understood. As a result, breast cancers have become an increasingly significant problem in the evaluation and management of patients.
The cDNA microarray technology allows us the parallel analysis of the expression of thousands of genes to address complex questions in tumor biology (1). Systematic investigation of expression pattern of thousands of genes in breast cancers using cDNA microarray, and their correlation to specific features of phenotypic variation, might provide the basis for an improved taxonomy of breast cancer (2-9). The ability to use appropriate profiles of gene expression could add enormous detail to the few known biological attributes in tumor characterization.
Elevated levels of β-catenin have been associated with poor prognosis in breast cancer (10). However, the mechanisms by which β-catenin confers the more aggressive biological behavior are poorly understood in breast cancer. β-catenin is a multifunctional protein that plays an essential role in the transduction of Wnt signaling and a component of the cadherin cell adhesion complex (11, 12). In the absence of Wnt signaling, the cytoplasmic level of β-catenin is kept low through interaction with a protein complex comprised of adenomatous polyposis coli (APC), Axin, protein phosphatase 2A (PP2A), and glycogen synthase kinase 3β (GSK3β), where β-catenin is phosphorylated by GSK3β (13, 14). Activation of Wnt signaing leads to the inactivation of GSK3β resulting in accumulation of cytoplasmic β-catenin. At elevated cytoplasmic levels, β-catenin translocates to the nucleus and cooperates with a member of the T cell factor (Tcf)/lymphocyte enhancer binding factor (Lef) family of high-mobility group box transcription factors to activate expression of target genes (13).
In this study, cDNA microarray analysis of approximately 4,600 cDNAs was performed to examine the gene expression profile in breast cancer on a more global scale. Immunohistochemical study was used to further demonstrate the specificity of the cDNA microarray results and to confirm the differential expression genes that were known to have important role in β-catenin regulation.
Ten pairs of breast cancer tissues and corresponding adjacent noncancerous breast tissues were obtained from 10 female patients who underwent lumpectomy or mastectomy at Dong-A University Medical Center between August 2001 and February 2002. Informed consent was obtained from patients to use their operated specimens and clinicopathological data for research purposes. The samples were anonymized before study. A pathologist dissected tissue samples from surgical specimens with special care for minimal contamination between cancer and noncancerous tissues and samples were immediately frozen in liquid nitrogen and stored at -80℃. Total RNA was isolated from the frozen tissue by using TRI REAGENT (Molecular Research Center, Inc., OH, U.S.A.). The remaining parts of specimens were fixed with 10% buffered formalin and the paraffin sections were stained with hematoxylin and eosin. Histological classifications were determined by two pathologists in each case. When there was a discrepancy in interpretation of pathologic grade between the two observers, the hig- her grade was selected as representative grade.
These clinical and histopathological features were summarized in Table 1.
Ten pairs of breast cancer and corresponding adjacent non-cancerous breast tissues were analyzed by MAGIC™ microarray (Macrogen, Inc., Korea). This array contains 4,600 cDNAs, isolated from cDNA library of human breast cancer, and is composed of 4,370 known genes and 230 unknown genes (EST). For synthesizing double-stranded cDNA probe, labeling kit provided by Macrogen was used. Microarray hybridization was performed according to manufacturer's instructions. Briefly, reverse transcription was performed on 30-50 µg of total RNAs for 1 hr at 42℃ with a T7-oligo (dT) 24 primer and Superscript II reverse transcriptase (Life Technologies, Inc., Rockville, MD, U.S.A.). Second-strand cDNA synthesis was performed for 2 hr at 16℃ with Escherichia coli DNA polymerase I, DNA ligase, and RNAse H (Life Technologies, Inc., Rockville, MD, U.S.A.), followed by incubation in solulion containing 50 mmol/L NaOH/ 0.1 mmol/L EDTA for 10 min at 65℃, which caused degradation of the rRNA and tRNA. After phenolchloroform extraction in vitro, transcription was performed for 6 hr at 37℃ using Cy5UTP and Cy3CTP, and a T7-Megascript kit (Ambion, Inc., Austin, TX, U.S.A.), and then fragmented for 35 min at 95℃. Hybridization (at 62℃ for 16 hr) with the labeled cDNA in 6 mL of hybridization solution (6× SSC, 0.2% SDS, 5× Denhardt solution, 1 mg/mL salmon sperm DNA) was followed by stringent washing (0.05× SSC, 0.2% SDS, at 58℃ for 1 hr).
Images were analyzed by using the ImaGene™ analysis software (BioDiscovery, Inc., Marina del Rey, CA, U.S.A.). Spots showing no signal, or obvious defects, were excluded from the analysis. The local background was subtracted from the remaining spots and the ratios of net fluorescence from the Cy5-specific channel to the net fluorescence from the Cy3-specific channel were calculated for each spot. For this purpose, we determined transcript abundance using a custom algorithm, and the data set was trimmed of genes expressed at extremely low levels; that is, a gene was excluded if its 70 percentile value was less than 0.2 in 2 [log (tumor/normal)] value. The results represented RNA expression in cancer relative to the corresponding noncancerous breast tissue.
A two-way pairwise average-linkage cluster analysis as an unsupervised clustering was applied. The cluster analysis was performed with Cluster 2.11, and the resulting expression map was visualized with TreeView 1.5. A set of microarray containing 4,600 cDNAs in duplicate was hybridized with a mixture of Cy3-labeled cDNA probes corresponding to cancer tissues and Cy5-labeled cDNA probes corresponding to noncancerous tissues. To identify genes with significantly different expressions in breast cancer and noncancerous tissues, rational Cy3/Cy5 cut off values were determined. The differentially expressed genes, identified from cDNA microarray comparisons, were assigned to a modification of the NCBI Clusters of Orthologous Gene classification by searching the OMIM and PubMed databases (15). The genes were then assigned to 10 known functions (cell cycle regulator, transcription, oncogene/tumor suppressor gene, metabolism, cell structure/motility, signal transduction, gene/protein expression, DNA repair, angiogenesis, and immunology) and a miscellaneous category.
Single-stranded cDNA was synthesized with oligo (dT) primer in a 20-µL reaction, from 5 µg of total SuperScript Preamplification System for First Strand cDNA Synthesis System (Life Technologies, Inc., Rockville, MD, U.S.A.) and diluted up to final volume of 80 µL. PCR was then performed with 1 µL of cDNA, for 1 cycle of 94℃ for 2 min, followed by 25 cycles of 94℃ for 30 sec, 55℃ for 30 sec, and 72℃ for 1 min, using a gene-specific primer and Taq polymerase.
Primers were as follows; U76248 (Siah) sense, 5'-AAGCTGTGATGTCCCATCTC-3', antisense, 5'-AGTCTGTAGGCAAAGTTCTC-3', AF009674 (Axin) sense, 5'-TGAGAACTCCAGACCGTTGT-3', antisense, 5'-TGCACATACCTCTGCTTGGA-3', D79995 sense, 5'-GGATGACGTGATACTCAATG-3', antisense, 5'-GGTAGAAAGGCTGGGCTCTT-3', X06825 (Tropomyosin 2β) sense, 5'-CTGGTGATCCTGGAAGGAGA-3', antisense, 5'-GCTTCTCCTCCAACAGTTTG-3'.
Amplification of the correct target DNA was confirmed by mobility of gel electrophoresis and sequencing after subcloning into pGEM-T easy vector (Promega, Madison, WI, U.S.A.). β-actin was used as an internal control to confirm equal amount between the templates.
Paraffin-embedded 5 µm-thick breast tumor sections were analyzed for β-catenin expression by immunohistochemical staining. Immunohistochemical staining was performed using monoclonal mouse anti-(human Ig) antibody directed against β-catenin (1:30, Zymed, CA, U.S.A.). Deparaffinization of all sections was performed through a series of xylene baths and rehydration was performed through graded alcohols. To enhance the immunoreactivity, microwave antigen retrieval at 750 W for 30 min in citrate buffer (pH 6.0) was performed. After blocking the endogenous peroxidase activity with 5% hydrogen peroxidase for 10 min, the primary antibody incubation for β-catenin was performed at room temperature for 100 min. Detection of the immunoreactive staining was carried out by the avidin-biotin-peroxidase complex method using Histostain-plus kit (Zymed, CA, U.S.A.). The antigen-antibody reaction was visualized using 3-amino-9-ethylcarbazole as a chromogen. The Mayer's hematoxylin counterstain was performed.
Immunohistochemical staining for β-catenin expression was divided into two groups according to their immunohistochemical patterns by a pathologist: normal membranous pattern seen in non-neoplastic epithelium and aberrantly accumulated pattern showing nuclear and/or cytoplasmic staining with or without membranous staining.
We categorized ten pairs of breast cancer by comparing the global gene expression patterns of cancer and noncancerous tissue after data trimming. Of the 4,600 genes, a total of 1,084 genes was left after data trimming. By unsupervised hierarchical clustering based on 1,084 genes, the cancer and non-cancerous tissues were successfully distinguished (data not shown). However, this algorithm using most of the genes on the array would not classify samples among cancer tissues by subgroups associated with clinical and histopathological features including age, tumor size, status of axillary lymph nodes, histologic type of the tumor, pathologic grade, and hormone-receptor status.
When cut off values were set to 2.0, for the ratio between the cancerous and noncancerous tissues, 106 (of which 55 were known) and 49 (of which 28 were known) genes or cDNA were up- or down-regulated, respectively, in >60% of the breast cancer samples. These genes were classified in terms of 10 functions (cell cycle regulator, transcription, oncogene/tumor suppressor gene, metabolism, cell structure/motility, signal transduction, gene/protein expression, DNA repair, angiogenesis, and immunology), as shown in Table 2. In the cancerous tissues, the genes related to cell cycle regulator, transcription, metabolism, cell structure/motility and signal transduction were mostly up-regulated. We identified consistent results of several genes that had some roles in cancer biology previously suggested by other workers, including pituitary tumor-transforming gene (PTTG) (16), histone acetyltransferase (HAT) (17), c-src tyrosine kinase (CSK) (18), and insulin-like growth factor binding protein (7).
To verify the reproducibility of these gene lists, we performed semiquantitative RT-PCR using the same RNA used in the microarray analysis. Ten pairs of the cancerous and corresponding noncancerous tissue samples were analyzed using four randomly selected genes; U76248 (Siah), AF009674 (Axin), D79995, and X06825 (Tropomysin 2β). Of these genes, two genes associated with β-catenin regulation, Siah, the mammalian homologue of Drosophila seven in absentia, and Axin, were included. The results of the breast cancer genes found from the semiquantitative RT-PCR analyses were mostly consistent with those from the microarray studies (Fig. 1).
When we reviewed genes that were differentially expressed in cancer and noncancerous tissue (Table 2), we could readily identify genes associated with β-catenin regulation. Three cancerous tissues (case 5-7) showing immunohistochemically aberrant accumulation of β-catenin revealed down-regulation of Siah and Axin genes and up-regulation of Wnt and c-myc genes. Of the remaining seven cancerous tissues without aberrant β-catenin expression, four cases (case 1, 2, 8, 9) showed that the results of the expression of these genes were vice versa and remaining three cases (case 3, 4, 10) showed inconsistent findings (Fig. 2, Table 3).
We have globally analyzed gene expression of breast cancer and noncancerous tissues to elucidate the characteristic changes associated with the carcinogenesis and progression in breast cancer. The cancer and noncancerous tissues were distinguished by gene expression profiling. In the cancer tissues, genes related to cell cycle, transcription, metabolism, cell structure/motility and signal transduction were mostly up-regulated in this study. We categorized ten pairs of breast cancer by comparing the global gene expression patterns of cancer and noncancerous tissue after data trimming. In this study, genes with expression ratios that varied at least 2-fold between the cancer and noncancerous tissues in 60% of the samples were selected. Although it resulted in the loss of some information, trimming in this manner decreased the possibility that the clustering algorithm would be strongly influenced by genes with little or no expression.
Most of the genes associated with the cell cycle were up-regulated, and examplified by the cyclin-dependent kinase 6 (CDK6) and PTTG. CDK6 is considered as a cdc-2 related kinase, which can play roles in the regulation of the mammalian cell cycle. The cyclin D/CDK6 complex phosphorylates the retinoblastoma protein. Under-phosphorylated pRb binds to the E2F family of transcription factors. Phosphorylation of the pRb unshackles the E2F proteins, which in turn activates the transcription of several genes whose products are essential for progression through the S phase (19). Because the state of the pRb phosphorylation is crucial in determining the cell cycle progression, the up-regulation of CDK6 in cancer tissues was not unexpected. PTTG has been identified as a key protein in mitotic regulation (20) and is highly expressed in pituitary tumor (21) and other neoplasms (22, 23). PTTG protein has been recognized as a mammalian securin protein that maintains binding of sister chromatids during mitosis. Overexpression of a nondegradable PTTG disrupts sister chromatid separation. PTTG overexpression results in genetic instability (24). Puri et al. (25) revealed a high level of expression of PTTG1 mRNA in both seminomatous and non-seminomatous testicular tumors, epithelial cell, sex-cord stromal cell, and germ cell tumors of the ovary, and in situ and invasive ductal carcinoma of the breast. In this study of breast cancer, PTTG was up-regulated in the cancerous tissue, which agrees with the results from a previous study (25) and suggests a certain role in tumorigenesis of breast cancer. On the other hand, the HAT directly links chromatin modification to genetic activation and has also important activities in many cellular processes including cell cycle progression, transcription and DNA replication (17). Developmental aberrations in mice and certain human cancers are associated with HAT mutations, further highlighting the important of these enzymes to normal cell growth and differentiation (26). In our study, this gene was down-regulated in cancer tissue which is consistent with the mechanism underlying tumor formation of mutations of HAT genes involved with the regulation and control of the cell cycle.
The loss of tumor suppressor gene is a key event in many, possibly all, human tumors, as the physiological function of these genes are in the regulation of normal cell growth including the cell cycle and nuclear transcription. We have identified a tumor suppressor gene that has previously been proposed by other research (18). The CSK has been shown to down-regulate the tyrosine kinase activity of the c-src oncoprotein through tyrosine phosphorylation of the c-src carboxyl terminus. The CSK gene could therefore potentially function as an antioncogene. Myoui et al. (27) examined the role of CSK in bone metastasis using an animal model with an inoculation of breast cancer cells and reported that CSK was an essential molecule for bone resorption by osteoclasts, which were central players in osteolytic bone metastases. They suggested the notion that CSK was a potential target molecule for designing novel therapeutic interventions, especially for bone metastases in breast cancer. In our study, CSK was down-regulated in cancer tissue of seven cases, which suggested that they needed to closer follow-up for bone metastasis. Identification of such a target may improve the efficiency of developing therapeutics for breast cancer.
cDNA microarray provided a powerful alternative with an unprecedented view scope in monitoring of the gene expression levels and led to discoveries of regulatory pathways involved in complicated biological processes. Differences in gene expression are likely to explain the phenotype variation. We used it to answer the specific question whether could be distinguished by their gene expression profiles and whether these could be related to differences in biological effects. Lin et al. (10) reported that high β-catenin activity significantly correlated with poor prognosis of the patients and was a strong and independent prognostic factor in breast cancer. Lim et al. (28) reported that aberrant expression of β-catenin was identified in 79% of breast cancer and there was correlation of aberrant expression of β-catenin with lymph node metastasis, survival rate, and survival length in Korean patients with breast cancer. β-catenin is an executor of Wnt signaling. Components of Wnt signaling pathway include proto-oncogene products, such as β-catenin, tumor suppressor proteins, such as APC, and Axin. It has been reported that Wnt genes are sometimes overexpressed in human breast cancer and there is a growing evidence that downstream components of the Wnt signaling pathway are activated in a significant proportion of breast tumors (29). Axin, a negative regulator of this pathway, promotes phoasphorylation of serine/threonine in exon 3 of β-catenin by forming a complex with APC and GSK-3β. Phosphorylated β-catenin is quickly degraded via an ubiquitin-proteasome pathway in the cytoplasm. Upon Wnt signaling, phosphorylation of β-catenin functions as a transcriptional regulator in the nucleus coupled with molecules of the Tcf/Lef family (13, 14). Siah is the mammalian homolog of Drosophila seven in absentia, required for formation of the R7 photoreceptor cells during eye development (30). Siah-1 mediated downregulation of β-catenin was induced by p53, providing a link between p53 and cell cycle regulation through a novel β-catenin degradation pathway (31). Our study indicated that β-catenin can be involved in breast cancer formation and/or progression and may serve as a target for breast cancer therapy, if we identified gene respect to regulatory pathways involved in β-catenin expression. In this study, three cancer tissues showing immunohistochemically aberrant accumulation of β-catenin in the nucleus and/or cytoplasm revealed down-regulation of Siah and Axin genes and up-regulation of Wnt and c-myc genes. Of the remaining seven cancer tissues without aberrant β-catenin expression, four cases showed that the results of these genes expression were vice versa. Although the remaining three cases showed inconsistent findings, our study revealed consistent findings of the mechanism of β-catenin expression in tumorigenesis of breast cancer. However, because the cases studied had short follow-up periods, we cannot determine the relation between β-catenin expression and prognostic significance.
We have analyzed whole cancer tissues in this study instead of focusing only on cancer cells to better describe the entire aspect of breast cancer, because breast cancer tissues as other cancers generally contain multiple nonepithelial cell types such as fibroblast, smooth muscle cell, endothelial cell, infiltrating lymphocyte, and macrophage. Recent advances in cancer research have revealed the relevance of epithelial-stromal interaction including extracellular matrices, matrix metalloproteinases, and angiogenic factors in cancer progression (32). Accordingly, targets of cancer therapeutics have been recently extended from molecules of cancer origin to molecules to stroma origin such as those related to angiogenesis and matrix remodeling (33, 34). As this study, it was based on whole tissue samples, the list of genes up-regulated in cancer tissues contained and may still contain many genes for stroma. With further functional study, potentially therapeutic target molecules of stroma might be identified.
These results provide not only a new molecular basis for understanding biological properties of breast cancer, but also useful resources for future development of therapeutic targets for breast cancer. To investigate the molecular understanding of regulatory pathways involved in the complicated carcinogenic processes, especially associated with the expression of β-catenin, a downstream effector of Wnt-mediated tumorigenesis in breast cancer, was also suggested. The genes suggested by this analysis may serve as candidates for more detailed examination by using a larger number of clinical samples and verification at the protein level.
Figures and Tables
Fig. 1
The results of semiquantitative RT-PCR for the 4 tested genes in the 10 breast cancer tissues and the noncancerous tissues, consisting with those from the microarrary studies. N, noncancerous tissue; T, tumor tissue; A, U76248 (Siah); B, AF009674 (Axin); C, D79995; D, X06825 (Tropomyosin 2β).
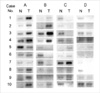
Fig. 2
Immunohistochemical stain for β-catenin in breast cancer. (A) Membranous pattern from case 1 (×200). (B) Aberrantly accumulated pattern in the nucleus and cytoplasm from case 5 (×400).
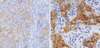
Table 1
Clinicopathologic summary of the breast cancer patients

Pathologic grade, histologic grade by Elston & Ellis method (35); LN, axillary lymph node metastasis; ER, estrogen receptor; PR, progesterone receptor; ca, carcinoma.
ACKNOWLEDGEMENT
This work was supported by the Korea Science and Engineering Foundation (KOSEF) through the MRCCMT at the Dong-A University.
References
1. Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996. 6:639–645.

2. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001. 98:10869–10874.

3. van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002. 415:530–536.
4. Jenssen TK, Kuo WP, Stokke T, Hovig E. Associations between gene expressions in breast cancer and patient survival. Hum Genet. 2002. 111:411–420.
5. van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002. 347:1999–2009.

6. Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, West M, Nevins JR, Huang AT. Gene expression predictors of breast cancer outcomes. Lancet. 2003. 361:1590–1596.

7. Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001. 61:5979–5984.
8. Kim HS, Jung JH, Park HY, Lee YH, Chung EJ, Kim MK, Kim JC. Gene expression profile analysis of human breast cancer using cDNA microarrays. J Korean Breast Cancer Soc. 2003. 6:58–67.
9. Han W, Chung KW, Ahn SJ, Noh DY, Youn YK, Oh SK, Choe KJ. Gene expression profiles of primary breast cancer tissue using cDNA microarray. J Korean Breast Cancer Soc. 2002. 5:284–290.

10. Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. β-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA. 1999. 97:4262–4266.
12. Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis: a look outside the nucleus. Science. 2000. 287:1606–1609.
13. Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997. 275:1787–1790.
14. Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P. Association of the APC gene product with β-catenin. Science. 1993. 262:1731–1734.
15. Lam AK. Molecular biology of esophageal squamous cell carcinoma. Crit Rev Oncol Hematol. 2000. 33:71–90.

16. McCabe C. Genetic targets for the treatment of pituitary adenomas: focus on the pituitary tumor transforming gene. Curr Opin Pharmacol. 2001. 1:620–625.

17. Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002. 4:287–299.

18. Armstrong E, Cannizzaro L, Bergman M, Huebner K, Alitalo K. The c-src tyrosine kinase (CSK) gene, a potential antioncogene, localizes to human chromosome region 15q23-25. Cytogenet Cell Genet. 1992. 60:119–120.
20. Zhang X, Horwitz GA, Prezant TR, Valentini A, Nakashima M, Bronstein MD, Melmed S. Structure, expression, and function of human pituitary tumor-transforming gene (PTTG). Mol Endocrinol. 1999. 13:156–166.

21. Zhang X, Horwitz GA, Heaney AP, Nakashima M, Prezant TR, Bronstein MD, Melmed S. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J Clin Endocrinol Metab. 1999. 84:761–767.

22. Dominguez A, Ramos-Morales F, Romero F, Rios RM, Dreyfus F, Tortolero M, Pintor-Toro JA. hpttg, a human homologue of rat pttg, is overexpressed in hematopoietic neoplasms. Evidence for a transcriptional activation function of hPTTG. Oncogene. 1998. 17:2187–2193.

23. Saez C, Japon MA, Ramos-Morales F, Romero F, Segura DI, Tortolero M, Pintor-Toro JA. hpttg is over-expressed in pituitary adenomas and other primary epithelial neoplasias. Oncogene. 1999. 18:5473–5476.

24. Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999. 285:418–422.

25. Puri R, Tousson A, Chen L, Kakar SS. Molecular cloning of pituitary tumor transforming gene 1 from ovarian tumors and its expression in tumors. Cancer Lett. 2001. 163:131–139.

26. Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev. 2001. 11:155–161.

27. Myoui A, Nishimura R, Williams PJ, Hiraga T, Tamura D, Michigami T, Mundy GR, Yoneda T. C-SRC tyrosine kinase activity is associated with tumor colonization in bone and lung in an animal model of human breast cancer metastasis. Cancer Res. 2003. 63:5028–5033.
28. Lim SC, Lee MS. Significance of E-cadherin/beta-catenin complex and cyclin D1 in breast cancer. Oncol Rep. 2002. 9:915–928.
29. Brown AM. Wnt signaling in breast cancer: have we come full circle? Breast Cancer Res. 2001. 3:351–355.

30. Carthew RW, Rubin GM. Seven in absentia, a gene required for specification R7 cell fate in the Drosophila eye. Cell. 1990. 63:561–577.
31. Liu J, Stevens J, Rote CA, Yost HJ, Hu Y, Neufeld KL, White RL, Matsunami N. Siah-1 mediates a novel β-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell. 2001. 7:927–936.

32. Matrisian LM, Cunha GR, Mohla S. Epithelial-stromal interactions and tumor progression: meeting summary and future directions. Cancer Res. 2001. 61:3844–3846.
33. Curran S, Murray GI. Matrix metalloproteinase: molecular aspects of their roles in tumor invasion and metastasis. Eur J Cancer. 2000. 36:1621–1630.
34. Deplanque G, Harris AL. Anti-angiogenic agents: clinical trial design and therapies in development. Eur J Cancer. 2000. 36:1713–1724.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download