Abstract
Recurrent spontaneous abortion (RSA) defines as two or more consecutive losses at ≤20 weeks of gestation and affects an estimated 1 of every 100 couples wishing to have children. However, it remains a poorly understood phenomenon. Recent reports observed a significant association between highly skewed X chromosome and RSA, supporting that X chromosome inactivation might be an important and previously unknown cause of RSA. X-inactivation pattern, using polymeric X-linked women with idiopathic RSA and 80 control subjects with a single successful pregnancy and no history of spontaneous abortion. The ratio of heterozygotes was 68.2% (45/66) in women with RSA and 67.5% (54/80) in control group. Among 45 informative RSA cases, only 1 (2.2%) woman showed extreme skewed X inactivation (≥90%) and 4 (8.9%) had mild skewed inactivation (≥85%). In 54 heterozygous control subjects, 5 (9.3%) women showed extreme skewed X inactivation and 7 (13.0%) had mild one. The frequency of skewed X inactivation between RSA patients and control group was not significantly different (p>0.05). This finding suggests that skewed X chromosome be not associated with unexplained RSA patients.
Recurrent spontaneous abortion (RSA), which is defined as two or more consecutive pregnancy losses before 22 weeks of gestation, is a prevalent disorder. It has been estimated to occur in 1-2% of couples wishing to have children (1). Although known causes of RSA include anatomic (15%), infectious (1-2%), hormonal (20%), immunologic (20%), and genetic (2-5%) disorders, 37-79% of those couples will not receive an explanation for their pregnancy losses (1, 2).
Recently, the finding was reported that skewed X chromosome inactivation was increased in women with RSA (3-5). X chromosome inactivation is defined as one of the two X chromosomes in each somatic cell of healthy human females becomes inactivated very early in embryonic development. This inactivation occurs randomly, on both the maternal and paternal X chromosome. Several studies have suggested that X-linked mutations underlie between RSA and skewed X chromosome inactivation (6). This hypothesis proposes that male lethal X-linked mutations cause skewed inactivation in female carriers. Many X-linked syndromes have been described in which carrier females exhibit highly skewed X inactivation (7-9). Such mutations are lethal to hemizygous male offsprings who inherit them, resulting in increased frequencies of spontaneous abortion in carrier females (3, 10).
A methylation sensitive assay was used to distinguish the active X chromosome from inactive one. This method is based on the assessment of the differential methylation between active and inactive X chromosomes. The human androgen receptor (AR gene, HUMARA, Xq11-12) locus, where methylation of this region correlates with X inactivation has been amplified by a rapid PCR assay. The first exon of AR locus has been widely used and reported to have a very high heterozygosity rate (11, 12).
To investigate whether skewed X chromosome inactivation is the potential cause of RSA or not, we studied DNA methylation patterns of the X-linked AR gene in patients experienced at least two losses as compared with reproductive women.
Between November 2001 and February 2003, 66 Korean women were collected as having recurrent pregnancy loss as defined by having 2 or more consecutive abortion and having RSA of unknown cause. Subjects presented with no other phenotypic abnormalities and with each pregnancy documented by a positive result on serum hCG, ultrasound or pathology. Also, subjects and those husbands had a normal karyotype. The mean age of patients was 33.5±0.5 yr, with a range of 27-43 yr. The mean number of consecutive spontaneous abortion per patient was 2.9, with range of 2-12. Control group included 80 healthy Korean women who had given birth to more than one child and no history of spontaneous or elective abortion. The mean age of control females was 33.9±0.4 yr, with a range of 27-43 yr. No significant difference in age was observed between RSA women and control group (p=0.457) (Fig. 1).
Genomic DNAs were extracted from peripheral blood using Wizard Genomic DNA Purification Kit (Promega, U.S.A.). A 600 ng portion of DNA from each sample was digested at 37℃ overnight with 8 U HpaII in a total volume of 22 µL of buffer L (Roche, Germany; 10 mM Tris-HCl, 10 mM MgCl2, 1 mM dithioerythritol, pH 7.5). After digestion, restriction enzyme was heat-inactivated by incubation at 95℃ for 15 min. Both undigested and digested DNA samples were included in reaction buffer (final concentration: 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.8 mM MgCl2), 200 µM each of dNTPs, 10 pmol each of primers, and 2 µU Amplitaq Gold (Perkin-Elmer, U.S.A.) in a total volume of 20 L. The sequences of the primers were AR1; 5'- GCT GTG AAG GTT GCT GTT CCT CAT-3'and AR2; 5'-TCC AGA ATC TGT TCC AGA GCG TGC-3'(11). The samples were amplified for 35 cycles comprising of 30 sec at 95℃, 10 sec at 66℃, 30 sec at 72℃. Electrophoresis was carried out using 8-15% polyacrylamide gel (29:1 acrylamide:bisacrylamide). Gels were stained with ethidium bromide and photographed under ultraviolet light.
DNA from male control was used as a digestion control, since his X chromosome is always active and unmethylated. Undigested male DNA showed a single band, while digested male DNA showed no PCR products indicating complete digestion by HpaII (Fig. 2D).
To quantify the methylation of AR alleles, we analyzed by BIO-PROFIL V.97 softwere (Vilber Lourmat, France). Degree of skewing was measured in relation to the most intense allele, meaning the inactivated X chromosome. The intensity of the digested sample was compared to measurements from the undigested sample. The incidence of skewing X chromosome was certified as (Bd1/Bu1)/(Bd1/Bu1+Bd2/Bu2) where Bd1 represents the band intensity of the more intense HpaII-digested allele, Bd2 represent the band intensity of the fainter HpaII-digested allele, and Bu1 and Bu2 are the corresponding bands from the undigested samples (13).
If a sample was found to have ≥75% skewing, the analysis was repeated in order to verify the result and an average was used. When more than 90% of an individual's lymphocytes had the same X chromosome active, X chromosome inactivation pattern was considered to be "extreme skewed". When the ratio of the intensity was more than 85%, the X inactivation was considered to be "mild skewed".
Fisher's exact test and χ2 analysis were applied for statistical analyses of the proportion of skewed X chromosome inactivation in RSA patients and control population. Student t-test was used to compare the age distribution among patient cases and control subjects. p values less than 0.05 were considered to indicate statistical significance.
We have examined X chromosome inactivation patterns in lymphocytes from 66 female women with idiopathic RSA and 80 control females who had one or more children and no history of spontaneous or elective abortion. Fig. 2 showed the representative examples of results gained by PCR. RSA30 appeared an X inactivation ratio of 56.95%, meaning random X chromosome inactivation (Fig. 2A), RSA32 having skewing ratio of 88.61% was found to show mild skewed inactivation (Fig. 2B), RSA48 having one PCR band was uninformative (Fig 2C), and digested male control showed no PCR products indicating complete digestion by HpaII (Fig. 2D). Of the 66 women with RSA, 45 (45/66, 68.2%) undigested DNA samples showed two PCR bands at the AR STR sites. Fifty-four women (54/80, 67.5%) of 80 control females were informative. The distribution of X inactivation ratios for both patients and controls is shown in Fig. 3.
In 45 informative RSA women, one (1/45, 2.2%) case was found to have extreme skewed X chromosome inactivation, defined as ≥90% inactivation of one allele by quantitative measurement. Also, 4 (4/45, 8.9%) had mild inactivation defined as ≥85% skewed X inactivation. Among 23 RSA patients having two consecutive losses, none (0/23, 0%) was determined to have extremely X inactivation and 1 (1/23, 4.3%) woman had mild one. One (1/22, 4.5%) of 22 female having three or more consecutive losses showed extreme X inactivation and 3 (3/22, 13.6%) showed mild one. In contrast, 5 (5/54, 9.3%) of 54 heterozygous control females observed extreme skewed X inactivation and 7 (7/54, 13.0%) showed mild X inactivation (Table 1). The differences in the frequency of skewed X inactivation was not significant between RSA patients and control group (p=0.216, ≥90% skewed; p=0.521, ≥85%). This finding suggests that skewed X chromosome be not associated with unexplained RSA patients.
Recently, skewed X-chromosome inactivation has been more frequently observed in women with idiopathic RSA. Lanasa and colleagues (6) found that the 14% of women with unexplained RSA show highly skewed X inactivation, which suggests that they are carriers of X-linked recessive lethal traits. Furthermore, female patients with highly skewed X inactivation were shown a significant decrease in male children. Uehara et al. (5) reported the incidence of preferential X inactivation in the women with RSA was 16.7%, a value that was much higher than that of the control group. Sangha et al. (3) presented evidence that factors associated with extremely skewed X-chromosome inactivation account for a significant proportion (18%) of couple with RSA, and Lanasa et al. (4) reported similar results for women who experienced 2 or more spontaneous abortions. However, Beever et al. (14) had published that the difference was not statistically significant, although women (12%) with RSA showed greater X inactivation skewing than did control women (7%). Our study, examined the association between RSA and skewed X chromosome inactivation, found that differences in the incidence of skewed X inactivation was not statistically significant. Also, the frequency (9.3%) of extremely skewed X inactivation (≥90%) in our control females was higher than that (2.2%) in women with RSA.
The incidence of skewed X chromosome inactivation in normal females is controversial. Previous studies with the AR methylation assay have reported that 1-23% of healthy females have extreme skewed X chromosome (ratios, ≥90) (Table 2). With the same criterion, our study has found the incidence of extreme skewing to be 9.3% in normal females. Because the AR methylation assay detected extreme skewing particularly in elderly females, we considered the age difference between RSA women and control groups (15-17) (Table 2). Therefore, we concluded that skewed X inactivation in our RSA and control females was not caused by an age effect, as the age of RSA woman with extreme skewed inactivation is 27 yr and those in control group were 29, 31, 35, 36, 39 yr (mean age 32.8 yr).
The human androgen receptor encoded in the X-chromosome is a member of the steroid receptor superfamily and contains a highly polymorphic CAG repeat sequence within exon 1. The 5'CpG island of the AR gene is methylated on the inactive X chromosome and hypomethylated on the active X chromosome (11). A methylation sensitive assay of AR locus was used to distinguish the active X chromosome from inactive one. This assay is the currently available method that permits reproducible PCR-based, but requires that the restriction digestion of the unmethylated DNA is complete. Therefore, the use of only a single methylation-sensitive restriction enzyme in the AR assay may tend to underestimate the true degree of skewing in some individuals and may explain the comparatively low incidences of severe skewed ratios (17). We have observed an incidence of 9.3% in control women, and male control samples showed no PCR products, indicating complete digestion by restriction enzyme.
Interestingly, Lanasa et al. (4, 6) and Uehara et al. (5) analyzed RSA women having two or more consecutive pregnancy losses, while Beever et al. (14) and Sangha et al. (3) defined RSA as three or more consecutive pregnancy losses. Between two groups, the incidences of extremely skewed X inactivation were likely to be similar. Our data showed that frequency of skewed inactivation in RSA cases with three or more consecutive losses are higher than that in women with two or more abortions (≥90, 4.5% vs. 2.2%; ≥85, 13.6% vs. 8.9%), but difference between the two RSA groups is not statistically significant (≥90, p=1.0; ≥85, p=0.675) (Table 1).
In summary, our findings suggest that extremely skewed X chromosome inactivation between RSA patients and control group is not significantly different contrary to several previous reports, and is not caused by an age effect. Furthermore, it is necessary to clarify the mechanism and to compare X inactivation patterns in peripheral blood to those in tissue cells from the patients.
Figures and Tables
Fig. 1
Comparison of age patterns in patients with RSA and control females. No significant difference in age was observed between two groups (p=0.457)

Fig. 2
Representative example of PCR analysis in three cases with RSA by AR gene. (A) RSA30 (skewed 56.95%); (B) RSA32 (skewed 88.61%); (C) RSA48, showing homozygosity (not informative); (D) normal male. Lane - and + show PCR amplification before and after HpaII digestion, respectively.

Fig. 3
Frequency distribution of X inactivation in 45 heterozygous RSA cases and 54 control females. The skewing values are expressed as the percentage ratio of the predominantly inactive allele to the predominantly active allele and are arranged into 10% intervals.
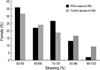
Table 1
Frequency of X inactivation in 45 heterozygous RSA cases and 54 control females
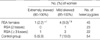
*p=0.216, Fisher's exact test, compared with control group having skewed ratio ≥90%; †p=0.521, χ2 test, compared with control group having skewed ratio ≥85%; ‡p=1.00, Fisher's exact test, compared with RSA (skewed ratio ≥90%) having three or more consecutive losses; §p=0.675, Fisher's exact test, compared with RSA (skewed ratio ≥85%) having three or more consecutive losses.
References
1. Stephenson MD. Frequency of factors associated with habitual abortion in 197 couples. Fertil Steril. 1996. 66:24–29.
2. Hatasaka HH. Recurrent miscarriage: epidemiologic factors, definitions, and incidence. Clin Obstet Gynecol. 1994. 37:625–634.

3. Sangha KK, Stephenson MD, Brown CJ, Robinson WP. Extremely skewed X-chromosome inactivation is increased in women with recurrent spontaneous abortion. Am J Hum Genet. 1999. 65:913–917.

4. Lanasa MC, Hogge WA, Kubik C, Blancato J, Hoffman EP. Highly skewed X-chromosome inactivation is associated with idiopathic recurrent spontaneous abortion. Am J Hum Genet. 1999. 65:252–254.

5. Uehara S, Hashiyada M, Sato K, Sato Y, Fujimori K, Okamura K. Preferential X-chromosome inactivation in women with idiopathic recurrent pregnancy loss. Fertil Steril. 2001. 76:908–914.

6. Lanasa MC, Hogge WA, Kubik CJ, Ness RB, Harger J, Nagel T, Prosen T, Markovic N, Hoffman EP. A novel X chromosome-linked genetic cause of recurrent spontaneous abortion. Am J Obstet Gynecol. 2001. 185:563–568.

7. Wengler G, Gorlin JB, Williamson JM, Rosen FS, Bing DH. Nonrandom inactivation of the X chromosome in early lineage hematopoietic cells in carriers of Wiskott-Aldrich syndrome. Blood. 1995. 85:2471–2477.

8. Naumova AK, Plenge RM, Bird LM, Leppert M, Morgan K, Willard HF, Sapienza C. Heritability of X chromosome-inactivation phenotype in a large family. Am J Hum Genet. 1996. 58:1111–1119.
9. Devriendt K, Matthijs G, Legius E, Schollen E, Blockmans D, van Geet C, Degreef H, Cassiman JJ, Fryns JP. Skewed X-chromosome inactivation in female carriers of dyskeratosis congenita. Am J Hum Genet. 1997. 60:581–587.
10. Pegoraro E, Whitaker J, Mowery-Rushton P, Surti U, Lanasa M, Hoffman EP. Familial skewed X inactivation: a molecular trait associated with high spontaneous-abortion rate maps to Xq28. Am J Hum Genet. 1997. 61:160–170.

11. Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992. 51:1229–1239.
12. Green AJ, Sepp T, Yates JR. Clonality of tuberous sclerosis harmatomas shown by non-random X-chromosome inactivation. Hum Genet. 1996. 97:240–243.

13. Lau AW, Brown CJ, Penaherrera M, Langlois S, Kalousek DK, Robinson WP. Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am J Hum Genet. 1997. 61:1353–1361.

14. Beever CL, Stephenson MD, Penaherrera MS, Jiang RH, Kalousek DK, Hayden M, Field L, Brown CJ, Robinson WP. Skewed X-chromosome inactivation is associated with trisomy in women ascertained on the basis of recurrent spontaneous abortion or chromosomally abnormal pregnancies. Am J Hum Genet. 2003. 72:399–407.

15. Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, Gilliland DG. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996. 88:59–65.

16. Kopp P, Jaggi R, Tobler A, Borisch B, Oestreicher M, Sabacan L, Jameson JL, Fey MF. Clonal X-inactivation analysis of human tumours using the human androgen receptor gene (HUMARA) polymorphism: a non-radioactive and semiquantitative strategy applicable to fresh and archival tissue. Mol Cell Probes. 1997. 11:217–228.

17. Sharp A, Robinson D, Jacobs P. Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet. 2000. 107:343–349.

18. Plenge RM, Stevenson RA, Lubs HA, Schwartz CE, Willard HF. Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet. 2002. 71:168–173.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download