Abstract
Purpose
Transcriptional silencing of tumor suppressor genes by aberrant methylation of CpG islands plays a crucial role in the development of human cancers. We comprehensively examined the methylation status of several tumor suppressor genes in osteosarcoma with a special focus on the RUNX3 gene.
Materials and Methods
Methylation-specific polymerase chain reaction (MSP) was performed for osteosarcoma tissues and their cell lines. MSP and RT-PCR for the RUNX3 gene were performed in the tumor-derived cell lines and the immortalized cell lines. The demethylating agent 5-aza-2' deoxycytidine was used in the SaOS-2 cell line to reverse the methylation status.
Results
Hypermethylation of the RUNX3 gene was observed in 60% (24 of 40) of the osteosarcoma tissues, whereas other tumor suppressor genes showed very low methylation. Thirteen of 30 (43%) tumor-derived cell lines, and U-2OS and SaOS-2 showed hypermethylation of the RUNX3 gene on MSPCR. However, RUNX3 was expressed in the SaOS-2 cell line, as determined by RT-PCR, and the expression was augmented by treatment with 5-aza-2' deoxycytidine.
Figures and Tables
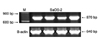
Fig. 2
Methylation of RUNX3 gene using methylation-specific PCR in osteosarcoma cell lines. (A) The results of 10 representative cell lines cultured from osteosarcoma tissues are shown. Numbers 2, 3, 7, 8 show methylated bands. (B) The results of 4 immortalized cell lines are shown. SaOS-2 and U-2OS showed methylation of RUNX3.

Fig. 3
Expression of RUNX3 in SaOS-2 cell line by RT-PCR. RUNX3 was expressed in SaOS-2 cell line. Treatment with Trichostatin A (TSA) down-regulated RUNX3 expression, which was reversed by co-treatment with 5-aza-2' deoxycytidine (5-aza-dC). Lane 1: control, lane 2: 5-aza-dC, lane 3: TSA, lane 4: 5-aza-dC+TSA.
References
1. Bacci G, Picci P, Ferrari S, et al. Primary chemotherapy and delayed surgery for nonmetastatic osteosarcoma of the extremities. Results in 164 patients preoperatively treated with high doses of methotrexate followed by cisplatin and doxorubicin. Cancer. 1993. 72:3227–3238.

2. Bangsow C, Rubins N, Glusman G, et al. The RUNX3 gene--sequence, structure and regulated expression. Gene. 2001. 279:221–232.
3. Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000. 25:315–319.

4. Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001. 61:3225–3229.
5. Gorlick R, Anderson P, Andrulis I, et al. Biology of childhood osteogenic sarcoma and potential targets for therapeutic development: meeting summary. Clin Cancer Res. 2003. 9:5442–5453.
6. Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997. 389:349–352.

7. Guo W, Wang X, Feng C. P53 gene abnormalities in osteosarcoma. Chin Med J (Engl). 1996. 109:752–755.
8. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996. 93:9821–9826.

9. Hou P, Ji M, Yang B, et al. Quantitative analysis of promoter hypermethylation in multiple genes in osteosarcoma. Cancer. 2006. 106:1602–1609.

10. Ito Y. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells. 1999. 4:685–696.

11. Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2'-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994. 91:11797–11801.

12. Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997. 57:808–811.
13. Kato N, Tamura G, Fukase M, Shibuya H, Motoyama T. Hypermethylation of the RUNX3 gene promoter in testicular yolk sac tumor of infants. Am J Pathol. 2003. 163:387–391.

14. Kim TY, Lee HJ, Hwang KS, et al. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest. 2004. 84:479–484.

15. Lewis IJ, Nooij MA, Whelan J, et al. Improvement in histologic response but not survival in osteosarcoma patients treated with intensified chemotherapy: a randomized phase III trial of the European Osteosarcoma Intergroup. J Natl Cancer Inst. 2007. 99:112–128.
16. MacLeod AR, Szyf M. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis. J Biol Chem. 1995. 270:8037–8043.

17. Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004. 9:422–441.

18. Matsubara H, Watanabe M, Imai T, et al. Involvement of extracellular signal-regulated kinase activation in human osteosarcoma cell resistance to the histone deacetylase inhibitor FK228 [(1S,4S,7Z,10S,16E,21R)-7-ethylidene-4,21-bis (propan-2-yl)-2-oxa-12,13-dit hia-5,8,20,23-tetraazabicyclo [8.7.6] tricos-16-ene-3,6,9,19,22-pentone]. J Pharmacol Exp Ther. 2009. 328:839–848.
19. Merlo A, Herman JG, Mao L, et al. 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995. 1:686–692.

20. Miller CW, Aslo A, Won A, Tan M, Lampkin B, Koeffler HP. Alterations of the p53, Rb and MDM2 genes in osteosarcoma. J Cancer Res Clin Oncol. 1996. 122:559–565.
21. Momparler RL. Molecular, cellular and animal pharmacology of 5-aza-2'-deoxycytidine. Pharmacol Ther. 1985. 30:287–299.

23. Peltonen K, Kiviharju TM, Järvinen PM, Ra R, Laiho M. Melanoma cell lines are susceptible to histone deacetylase inhibitor TSA provoked cell cycle arrest and apoptosis. Pigment Cell Res. 2005. 18:196–202.

24. Rini D, Calabi F. Identification and comparative analysis of a second runx3 promoter. Gene. 2001. 273:13–22.

25. Roh MS, Kim CW, Park BS, et al. Mechanism of histone deacetylase inhibitor Trichostatin A induced apoptosis in human osteosarcoma cells. Apoptosis. 2004. 9:583–589.

26. Romanski A, Bacic B, Bug G, et al. Use of a novel histone deacetylase inhibitor to induce apoptosis in cell lines of acute lymphoblastic leukemia. Haematologica. 2004. 89:419–426.
27. Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984. 81:6993–6997.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download