Abstract
Carbapenem-resistant Klebsiella pneumoniae isolates producing K. pneumoniae carbapenemases (KPC) were first reported in the USA in 2001, and since then, this infection has been reported in Europe, Israel, South America, and China. In Korea, the first KPC-2-producing K. pneumoniae sequence type (ST) 11 strain was detected in 2010. We report the case of a patient with a urinary tract infection caused by KPC-2-producing K. pneumoniae. This is the second report of a KPC-2-producing K. pneumoniae infection in Korea, but the multilocus sequence type was ST258. The KPC-2-producing isolate was resistant to all tested β-lactams (including imipenem and meropenem), amikacin, tobramycin, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole, but was susceptible to gentamicin, colistin, polymyxin B, and tigecycline. The KPC-2-producing isolate was negative to phenotypic extended-spectrum β-lactamase (ESBL) and AmpC detection tests and positive to modified Hodge test and carbapenemase inhibition test with aminophenylboronic acid.
The resistance of Klebsiella pneumoniae to carbapenems is mainly associated with acquired carbapenemases [1]. These carbapenemases can be Ambler class A (KPC, GES), class B (VIM, IMP), and class D (OXA-48) enzymes [2]. The most common class A carbapenemases in K. pneumoniae are the K. pneumoniae carbapenemases (KPCs) [3]. KPC-producing K. pneumoniae strains were first reported in 2001 in the USA [4] and dissemination has been reported in Europe, Israel, South America, and China [5-9]. The first KPC-2-producing K. pneumoniae strain in Korea was isolated from bronchial aspirates from a patient admitted to the intensive care unit (ICU) in 2010 [10]. KPC-producing K. pneumoniae also produce VIM or CTX-M, making it difficult to select appropriate antibiotics [11]. In addition, the mortality rate is significantly higher for patients with KPC-producing isolates than those with imipenem susceptible isolates [12]. In this report, we describe a case of infection with a KPC-2-producing K. pneumoniae isolate, sequence type (ST) 258 in Korea and various phenotypic methods for screening and confirmation.
A 70-year-old woman was admitted to the Plastic Surgery (PS) department on October 5, 2010, with a 24-h history of fever and dizziness. She had a known history of unstable angina and diabetes mellitus (2001). In November 2009, she was admitted to the PS department for a skin flap operation (February, 2010) to treat a third-degree burn to the sacral area. Two months ago, a sore, approximately 10×10 cm in size, developed at the sacral area and progressed to osteomyelitis in the sacral bone. She had no recent travel history abroad. At admission, she was pale and febrile with a temperature of 38.2℃. An intermittent fever of 37.3-38.1℃ lasted until hospital day (HD) 5. Her blood pressure was 110/80 mmHg, her pulse was 78/min, and her respiratory rate was 20/min. A laboratory investigation at the time of admission revealed a peripheral white blood cell (WBC) count of 8,730/µL (73.5% neutrophils), a hemoglobin level of 8.6 g/dL, and a platelet count of 261,000/µL. Routine blood chemistry results were AST/ALT of 7/11 U/L, alkaline phosphatase of 77 U/L, blood urea nitrogen/creatinine of 23.4/1.42 mg/dL, and total protein/albumin of 5.6/2.9 g/dL. Erythrocyte sedimentation rate and C-reactive protein were both increased to 53 mm/hr and 220.03 mg/L, respectively. The urine was yellow and turbid and routine urinalysis revealed a positive WBC (3+), and positive protein (1+). Microscopic examination of urine revealed >60 WBCs and yeast organisms in a high power field. A chest radiograph showed right pleural thickening and a little collapse of the right lower lung. An abdomen and pelvic computed tomography showed signs of cystitis and fluid collection in both the abdomen and pleural cavity.
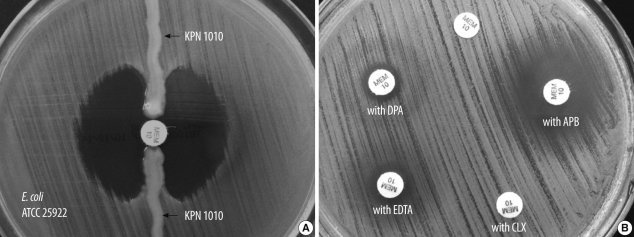
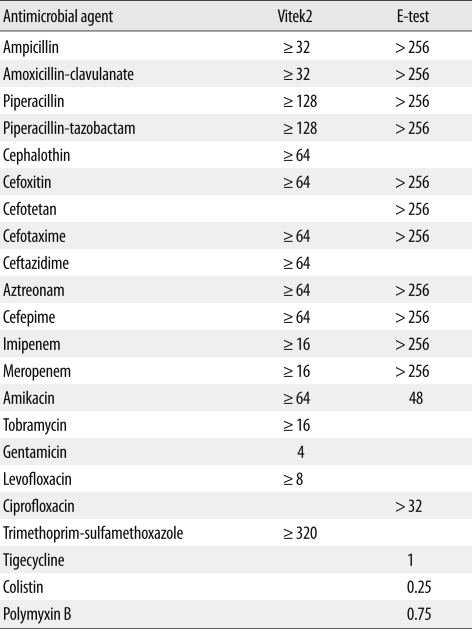
Two sets of blood culture bottles and a urine sample were taken for microbiologic study. Aerobic and anaerobic blood cultures were all negative after 5 days of incubation. In the urine culture, Candida albicans (8×104 CFU/mL) grew on a blood agar plate. On HD 32, the patient had a fever of 38.1℃. Two sets of blood culture bottles and a urine sample were collected again for culture study. The blood culture results were negative. Multidrug-resistant K. pneumoniae (KPN 1010, >105 CFU/mL) was isolated from the urine. Vitek2 GN and AST-N044 (bioMérieux, Marcy l'Étoile, France) were used for species identification and antimicrobial susceptibility test, respectively. With the exception of gentamicin, all susceptibility results showed high-level minimum inhibitory concentration (MIC) values. MICs were also assessed by E-test (bioMérieux). Most antibiotics were resistant and consistent with the MIC of Vitek2. However, MICs of tigecycline and colistin were 1 and 0.25 µg/mL, respectively (Table 1). The modified Hodge test [13] demonstrated strong positivity (Fig. 1A) but AmpC and ESBL phenotypic tests [14] were negative. Carbapenemase inhibition tests were performed for discrimination of carbapenemases. Briefly, meropenem disks (Becton-Dickinson, Cockeysville, MD, USA) were supplemented with 10 µL of 4 different β-lactamase inhibitors: 60 mg/mL aminophenylboronic acid (APB; Sigma St. Louis, MO, USA), 75 mg/mL cloxacillin (Sigma), 100 mg/mL dipicolinic acid (DPA; Sigma), and 0.2 M ethylenediaminetetraacetic acid (EDTA; Sigma). A 0.5 McFarland inoculum was prepared and spread on Mueller-Hinton agar plates (Becton-Dickinson). Five disks were placed on each plate: meropenem, meropenem+APB, meropenem+cloxacillin, meropenem+DPA, meropenem+EDTA. A positive response was achieved when there was a greater than 5 mm increase of the inhibition zone diameter around disks containing β-lactamase inhibitors, as compared with the meropenem disk alone [15]. The positive result was seen only with APB (Fig. 1B).
PCR and DNA sequencing were performed with primers specific for the blaVIM, blaIMP, blaSIM, blaNDM, and blaKPC genes [16-18]. We found only the blaKPC-2 gene. Multilocus sequence typing (MLST) with 7 housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB) was performed [19]. The MLST showed that the isolate belonged to the epidemic clone ST258. Initially, the patient was treated with ertapenem (1 g once a day). After multidrug-resistant (MDR) K. pneumoniae was isolated from urine and the sacral sore wound, tigecycline (50 mg twice a day) was administrated for 14 days. However, neither pyuria nor the wound subsided, and MDR K. pneumoniae was repeatedly isolated. Lastly, the patient was treated with colistin (150 mg once a day), but clinical improvement was not observed and kidney function was declining. She was managed in ICU, but expired on the 90th HD due to septic shock and multiorgan failure.
The most common mechanism of carbapenem-resistant K. pneumoniae in Korea is ESBL and/or AmpC β-lactamase plus porin loss [20]. Only a single case of KPC-2-producing strain has been described in Korea and the MLST type was ST11 [10]. This isolate showed an MDR pattern to various antibiotics, including colistin. The previous patient was treated with colistin, but the prognosis was also poor. This is the second report of a KPC-2-producing K. pneumoniae in Korea. The isolate was not associated with travel and the MLST type was ST258. ST258 accounts for 70% of KPC-producing K. pneumoniae in the USA [21]. Isolates of ST258 have also been identified in Europe and Israel, indicating an international spread of ST258 among KPC-producing K. pneumoniae [6, 7].
Most KPC-producing K. pneumoniae have been associated with other β-lactamase genes, such as the widespread ESBL gene blaCTX-M [22]. The previous KPC-2-producing K. pneumoniae isolate in Korea also contained blaCTX-M-15 [10]. However, the KPN 1010 isolate contained neither ESBL nor AmpC gene in this study.
The KPN 1010 isolate was susceptible to gentamicin, colistin, polymyxin B, and tigecycline, but the previous Korean isolate was nonsusceptible to gentamicin, colistin, polymyxin B, and tigecycline [10].
A modified Hodge test accurately detects KPC, but is not able to discriminate from other carbapenemases [23]. A carbapenemase inhibition test, comprising a meropenem disk, and meropenem disks supplemented with APB (for detection of class A carbapenemases), cloxacillin (for detection of AmpC β-lactamases plus porin loss), DPA or EDTA (for detection of class B metallo-carbapenemases) accurately distinguishes between several different mechanisms mediating reduced susceptibility to carbapenems in Enterobacteriaceae [15]. The isolate was positive to APB and negative to cloxacillin, DPA, and EDTA, suggesting a class A carbapenemase producing isolate.
References
1. Nordmann P, Poirel L. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect. 2002; 8:321–331. PMID: 12084099.

2. Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007; 20:440–458. PMID: 17630334.

3. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009; 9:228–236. PMID: 19324295.
4. Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001; 45:1151–1161. PMID: 11257029.
5. Giakkoupi P, Pappa O, Polemis M, Vatopoulos AC, Miriagou V, Zioga A, et al. Emerging Klebsiella pneumoniae isolates coproducing KPC-2 and VIM-1 carbapenemases. Antimicrob Agents Chemother. 2009; 53:4048–4050. PMID: 19581459.
6. Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, et al. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother. 2009; 53:818–820. PMID: 19029323.
7. Samuelsen Ø, Naseer U, Tofteland S, Skutlaberg DH, Onken A, Hjetland R, et al. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J Antimicrob Chemother. 2009; 63:654–658. PMID: 19218573.
8. Villegas MV, Lolans K, Correa A, Suarez CJ, Lopez JA, Vallejo M, et al. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother. 2006; 50:2880–2882. PMID: 16870793.
9. Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007; 51:763–765. PMID: 17145797.
10. Rhee JY, Park YK, Shin JY, Choi JY, Lee MY, Peck KR, et al. KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob Agents Chemother. 2010; 54:2278–2279. PMID: 20211897.
11. Pournaras S, Poulou A, Voulgari E, Vrioni G, Kristo I, Tsakris A. Detection of the new metallo-β-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae. J Antimicrob Chemother. 2010; 65:1604–1607. PMID: 20522444.
12. Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother. 2008; 52:1413–1418. PMID: 18227191.
13. Lee K, Kim CK, Yong D, Jeong SH, Yum JH, Seo YH, et al. Improved performance of the modified Hodge test with MacConkey agar for screening carbapenemase-producing Gram-negative bacilli. J Microbiol Methods. 2010; 83:149–152. PMID: 20801167.

14. Song W, Jeong SH, Kim JS, Kim HS, Shin DH, Roh KH, et al. Use of boronic acid disk methods to detect the combined expression of plasmid-mediated AmpC β-lactamases and extended-spectrum β-lactamases in clinical isolates of Klebsiella spp., Salmonella spp., and Proteus mirabilis. Diagn Microbiol Infect Dis. 2007; 57:315–318. PMID: 17174510.
15. Giske CG, Gezelius L, Samuelsen Ø, Warner M, Sundsfjord A, Woodford N. A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin Microbiol Infect. 2011; 17:552–556. PMID: 20597925.
16. Patzer JA, Walsh TR, Weeks J, Dzierzanowska D, Toleman MA. Emergence and persistence of integron structures harbouring VIM genes in the Children's Memorial Health Institute, Warsaw, Poland, 1998-2006. J Antimicrob Chemother. 2009; 63:269–273. PMID: 19095681.

17. Smith Moland E, Hanson ND, Herrera VL, Black JA, Lockhart TJ, Hossain A, et al. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J Antimicrob Chemother. 2003; 51:711–714. PMID: 12615876.
18. Zarfel G, Hoenigl M, Leitner E, Salzer HJ, Feierl G, Masoud L, et al. Emergence of New Delhi metallo-β-lactamase, Austria. Emerg Infect Dis. 2011; 17:129–130. PMID: 21192874.

19. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005; 43:4178–4182. PMID: 16081970.
20. Park YJ, Yu JK, Park KG, Park YG, Lee S, Kim SY, et al. Prevalence and contributing factors of nonsusceptibility to imipenem or meropenem in extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Escherichia coli. Diagn Microbiol Infect Dis. 2011; in press.
21. Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009; 53:3365–3370. PMID: 19506063.
22. Cai JC, Zhou HW, Zhang R, Chen GX. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli Isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother. 2008; 52:2014–2018. PMID: 18332176.
23. Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, et al. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007; 45:2723–2725. PMID: 17581941.
Fig. 1
Results obtained with a Modified Hodge test (A) and carbapenemase inhibition test (B) for carbapenem-resistant Klebsiella pneumoniae isolate (KPN 1010).
Abbreviations: MEM, meropenem; APB, aminophenylboronic acid; CLX, cloxacillin; EDTA, ethylenediaminetetraacetic acid; DPA, dipicolinic acid.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download